3DGK
 
 | |
3F5G
 
 | | Crystal structure of death associated protein kinase in complex with ADP and Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Death-associated protein kinase 1, MAGNESIUM ION | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-03 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3F5U
 
 | | Crystal structure of the death associated protein kinase in complex with AMPPNP and Mg2+ | | Descriptor: | Death-associated protein kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3CHT
 
 | |
3F9X
 
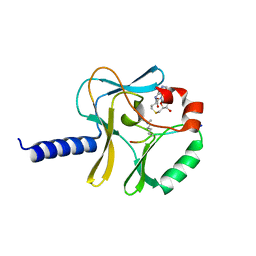 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20me2 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3FF1
 
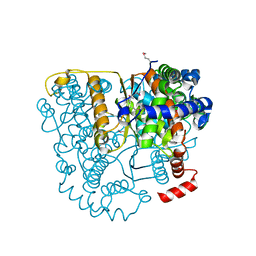 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3DFM
 
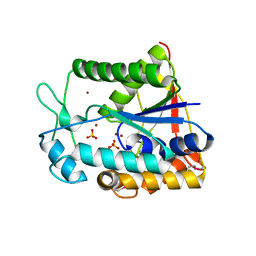 | |
3EDN
 
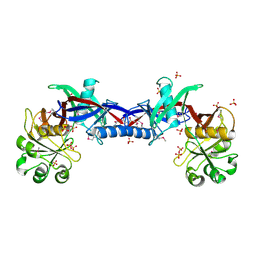 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | Descriptor: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
3GEU
 
 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
3EUQ
 
 | |
3EUT
 
 | |
3DFF
 
 | | The crystal structure of teicoplanin pseudoaglycone deacetylase Orf2 | | Descriptor: | GLYCEROL, TETRAETHYLENE GLYCOL, Teicoplanin pseudoaglycone deacetylases Orf2, ... | | Authors: | Zou, Y, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-06-11 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of lipoglycopeptide antibiotic deacetylases: implications for the biosynthesis of a40926 and teicoplanin.
Chem.Biol., 15, 2008
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
3ERP
 
 | | Structure of IDP01002, a putative oxidoreductase from and essential gene of Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Minasov, G, Evdokimova, E, Brunzelle, J.S, Kudritska, M, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-02 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl.Environ.Microbiol., 2017
|
|
5UWG
 
 | | The crystal structure of Fz4-CRD in complex with palmitoleic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, PALMITOLEIC ACID | | Authors: | Ke, J, DeBruine, Z.J, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Wnt5a promotes Frizzled-4 signalosome assembly by stabilizing cysteine-rich domain dimerization.
Genes Dev., 31, 2017
|
|
4LEU
 
 | | Crystal Structure of THA8-like protein from Arabidopsis thaliana | | Descriptor: | Pentatricopeptide repeat-containing protein At3g46870 | | Authors: | Ke, J, Chen, R.Z, Ban, T, Brunzelle, J.S, Gu, X, Kang, Y, Melcher, K, Zhu, J.K, Xu, H.E. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a PLS-class Pentatricopeptide Repeat Protein Provides Insights into Mechanism of RNA Recognition.
J.Biol.Chem., 288, 2013
|
|
5T0Q
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T0F
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3K29
 
 | | Structure of a putative YscO homolog CT670 from Chlamydia trachomatis | | Descriptor: | Putative uncharacterized protein | | Authors: | Lam, R, Singer, A, Skarina, T, Onopriyenko, O, Bochkarev, A, Brunzelle, J.S, Edwards, A.M, Anderson, W.F, Chirgadze, N.Y, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and protein-protein interaction studies on Chlamydia trachomatis protein CT670 (YscO Homolog).
J.Bacteriol., 192, 2010
|
|
