4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4UTT
 
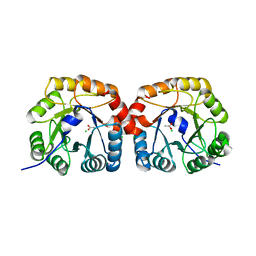 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | ACETATE ION, CHLORIDE ION, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway.
J.Biol.Chem., 289, 2014
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1GWE
 
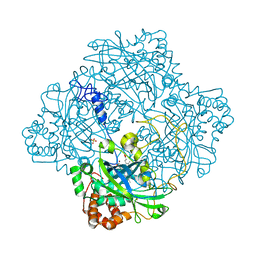 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GWH
 
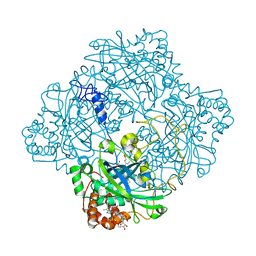 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase complexed with NADPH | | Descriptor: | ACETATE ION, Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1H4X
 
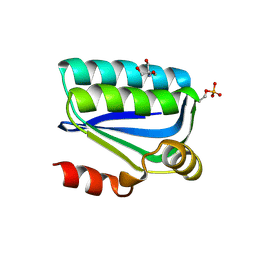 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1T9H
 
 | | The crystal structure of YloQ, a circularly permuted GTPase. | | Descriptor: | ACETATE ION, CALCIUM ION, Probable GTPase engC, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Cladiere, L, Antson, A.A, Isupov, M.N, Seror, S.J, Wilkinson, A.J. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
J.Mol.Biol., 340, 2004
|
|
4UTU
 
 | | Structural and biochemical characterization of the N- acetylmannosamine-6-phosphate 2-epimerase from Clostridium perfringens | | Descriptor: | CHLORIDE ION, N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE, N-acetylmannosamine-6-phosphate | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
3PVA
 
 | | PENICILLIN V ACYLASE FROM B. SPHAERICUS | | Descriptor: | PROTEIN (PENICILLIN V ACYLASE) | | Authors: | Suresh, C.G, Pundle, A.V, Rao, K.N, Sivaraman, H, Brannigan, J.A, Mcvey, C.E, Verma, C.S, Dauter, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members.
Nat.Struct.Biol., 6, 1999
|
|
4UFW
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 22) | | Descriptor: | 2-oxopentadecyl-CoA, 4-chloranyl-N-[2-(3-methoxyphenyl)ethanimidoyl]-2-piperidin-4-yloxy-benzamide, CHLORIDE ION, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
4UFX
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 19) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
4UFV
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 18) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
6H75
 
 | | SiaP A11N in complex with Neu5Ac (RT) | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6H76
 
 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
4PEL
 
 | | S1C mutant of Penicillin G acylase from Kluyvera citrophila | | Descriptor: | CALCIUM ION, Penicillin G acylase subunit alpha, Penicillin G acylase subunit beta | | Authors: | Ramasamy, S, Chand, D, Varshney, N.K, Brannigan, J.A, Wilkinson, A.J, Suresh, C.G. | | Deposit date: | 2014-04-24 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Penicillin G acylase
To Be Published
|
|
4PEM
 
 | | Crystal Structure of S1G mutant of Penicillin G Acylase from Kluyvera citrophila | | Descriptor: | CALCIUM ION, Penicillin G acylase Alpha, Penicillin G acylase Beta | | Authors: | Ramasamy, S, Varshney, N.K, Brannigan, J.A, Wilkinson, A.J, Suresh, C.G. | | Deposit date: | 2014-04-24 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S1G mutant of Penicillin G Acylase from Kluyvera citrophilla
To be Published
|
|
5G20
 
 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 19). | | Descriptor: | 6-(BENZYLOXY)-4-(ETHYLSULFANYL)-3-[(MORPHOLIN-4-YL), DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G1Z
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 1) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G21
 
 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 26). | | Descriptor: | ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G22
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 26) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
1B0N
 
 | | SINR PROTEIN/SINI PROTEIN COMPLEX | | Descriptor: | PROTEIN (SINI PROTEIN), PROTEIN (SINR PROTEIN), ZINC ION | | Authors: | Lewis, R.J, Brannigan, J.A, Offen, W.A, Smith, I, Wilkinson, A.J. | | Deposit date: | 1998-11-11 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex.
J.Mol.Biol., 283, 1998
|
|
2D30
 
 | | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution | | Descriptor: | ZINC ION, cytidine deaminase | | Authors: | Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Moroz, O.V, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-21 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution
To be Published
|
|
2BZB
 
 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-08-14 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2C8J
 
 | | CRYSTAL STRUCTURE OF ferrochelatase HemH-1 from Bacillus anthracis, str. Ames | | Descriptor: | FERROCHELATASE 1 | | Authors: | Muller, A, Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-12-05 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ferrochelatase Hemh-1 from Bacillus Anthracis, Str. Ames
To be Published
|
|
2BKX
 
 | |
