4ARE
 
 | | Crystal structure of the collagenase Unit of collagenase G from Clostridium histolyticum at 2.19 angstrom resolution. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, COLLAGENASE G, ... | | Authors: | Eckhard, U, Brandstetter, H. | | Deposit date: | 2012-04-23 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for Activity Regulation and Substrate Preference of Clostridial Collagenases G, H, and T.
J.Biol.Chem., 288, 2013
|
|
1XQY
 
 | | Crystal structure of F1-mutant S105A complex with PRO-LEU-GLY-GLY | | Descriptor: | PLGG, PROLINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XQX
 
 | | Crystal structure of F1-mutant S105A complex with PCK | | Descriptor: | PHENYLALANYLMETHYLCHLORIDE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRM
 
 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Ala-Phe | | Descriptor: | ALANINE, PHENYLALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRP
 
 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Pro-Leu-Gly-Gly | | Descriptor: | PLGG, PROLINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRN
 
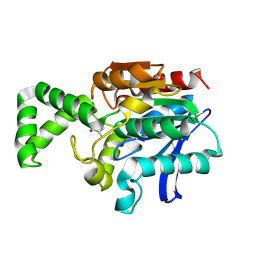 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Phe-Ala | | Descriptor: | ALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRR
 
 | | Crystal structure of active site F1-mutant E245Q soaked with peptide Pro-Pro | | Descriptor: | PROLINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XQV
 
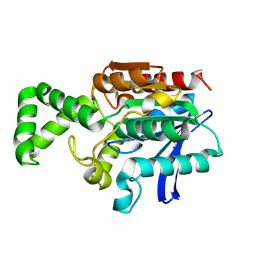 | | Crystal structure of inactive F1-mutant G37A | | Descriptor: | Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRL
 
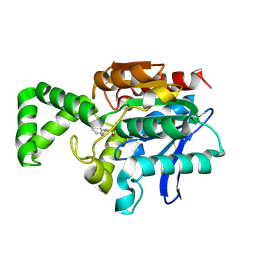 | | Crystal structure of active site F1-mutant Y205F complex with inhibitor PCK | | Descriptor: | (2R,3S)-3-AMINO-1-CHLORO-4-PHENYL-BUTAN-2-OL, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XQW
 
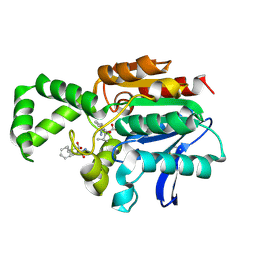 | | Crystal structure of F1-mutant S105A complex with PHE-LEU | | Descriptor: | LEUCINE, PHENYLALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRQ
 
 | | Crystal structure of active site F1-mutant E245Q soaked with peptide Phe-Leu | | Descriptor: | LEUCINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRO
 
 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Phe-Leu | | Descriptor: | LEUCINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1MT3
 
 | | Crystal Structure of the Tricorn Interacting Factor Selenomethionine-F1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
1Z1W
 
 | | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 394, 2005
|
|
1MTZ
 
 | | Crystal Structure of the Tricorn Interacting Factor F1 | | Descriptor: | Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
1MU0
 
 | | Crystal Structure of the Tricorn Interacting Factor F1 Complex with PCK | | Descriptor: | (2R,3S)-3-AMINO-1-CHLORO-4-PHENYL-BUTAN-2-OL, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
1ZRZ
 
 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
4BQD
 
 | | KD1 of human TFPI in complex with a synthetic peptide | | Descriptor: | GLYCEROL, PEPTIDE, TISSUE FACTOR PATHWAY INHIBITOR (LIPOPROTEIN-ASSOCIATED COAGULATION INHIBITOR) VARIANT | | Authors: | Griessner, A, Brandstetter, H. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Small Peptides Blocking Inhibition of Factor Xa and Tissue Factor-Factor Viia by Tissue Factor Pathway Inhibitor (Tfpi)
J.Biol.Chem., 289, 2014
|
|
4AR8
 
 | | Crystal structure of the peptidase domain of collagenase T from Clostridium tetani complexed with the peptidic inhibitor isoamyl- phosphonyl-Gly-Pro-Ala at 2.05 angstrom resolution. | | Descriptor: | CALCIUM ION, COLLAGENASE COLT, ISOAMYL-PHOSPHONYL-GLY-PRO-ALA, ... | | Authors: | Eckhard, U, Brandstetter, H. | | Deposit date: | 2012-04-21 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Activity Regulation and Substrate Preference of Clostridial Collagenases G, H, and T.
J.Biol.Chem., 288, 2013
|
|
4ARF
 
 | | CRYSTAL STRUCTURE OF THE PEPTIDASE DOMAIN OF COLLAGENASE H FROM CLOSTRIDIUM HISTOLYTICUM IN COMPLEX WITH THE PEPTIDIC INHIBITOR ISOAMYLPHOSPHONYL-GLY-PRO-ALA AT 1.77 ANGSTROM RESOLUTION. | | Descriptor: | CALCIUM ION, COLH PROTEIN, ISOAMYLPHOSPHONYL-GLY-PRO-ALA, ... | | Authors: | Eckhard, U, Brandstetter, H. | | Deposit date: | 2012-04-23 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis for Activity Regulation and Substrate Preference of Clostridial Collagenases G, H, and T.
J.Biol.Chem., 288, 2013
|
|
4AR1
 
 | |
4AR9
 
 | |
4AWB
 
 | | Crystal structure of active legumain in complex with AAN-CMK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEGUMAIN, MERCURY (II) ION, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic and Structural Studies on Legumain Explain its Zymogenicity, Distinct Activation Pathways, and Regulation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AW9
 
 | | Crystal structure of active legumain in complex with YVAD-CMK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-TYR-VAL-ALA-ASP-CHLOROMETHYLKETONE, LEGUMAIN, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Studies on Legumain Explain its Zymogenicity, Distinct Activation Pathways, and Regulation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BK7
 
 | |
