1ZGD
 
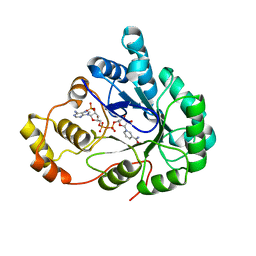 | | Chalcone Reductase Complexed With NADP+ at 1.7 Angstrom Resolution | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, chalcone reductase | | Authors: | Bomati, E.K, Austin, M.B, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis
J.Biol.Chem., 280, 2005
|
|
1EYP
 
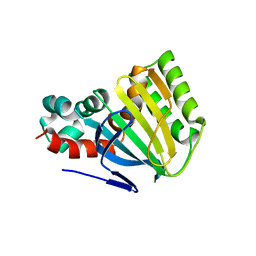 | | CHALCONE ISOMERASE | | Descriptor: | CHALCONE-FLAVONONE ISOMERASE 1 | | Authors: | Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-05-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase.
Nat.Struct.Biol., 7, 2000
|
|
4DOK
 
 | |
4DOO
 
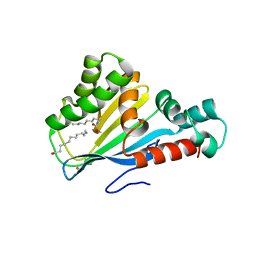 | | Crystal structure of Arabidopsis thaliana fatty-acid binding protein At3g63170 (AtFAP1) | | Descriptor: | Chalcone-flavanone isomerase family protein, LAURIC ACID, POTASSIUM ION | | Authors: | Noel, J.P, Pojer, F, Louie, G.V, Bowman, M.E. | | Deposit date: | 2012-02-09 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis.
Nature, 485, 2012
|
|
6MS8
 
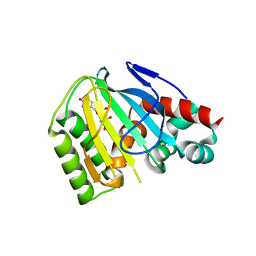 | | Crystal Structure of Chalcone Isomerase from Medicago Truncatula Complexed with (2S) Naringenin | | Descriptor: | Chalcone-flavonone isomerase family protein, NARINGENIN | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
8DLE
 
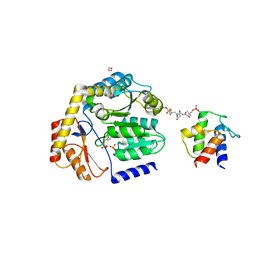 | | Crosslinked Crystal Structure of the 8-amino-7-oxonanoate synthase, BioF, and Benzene Sulfonyl Fluoride-crypto Acyl Carrier Protein, BSF-ACP | | Descriptor: | 1,2-ETHANEDIOL, 8-amino-7-oxononanoate synthase, Acyl carrier protein, ... | | Authors: | Chen, A, Davis, T.D, Louie, G.V, Bowman, M.E, Noel, J.P, Burkart, M.D. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing the Interface of Biotin and Fatty Acid Biosynthesis through SuFEx Probes.
J.Am.Chem.Soc., 146, 2024
|
|
1F8A
 
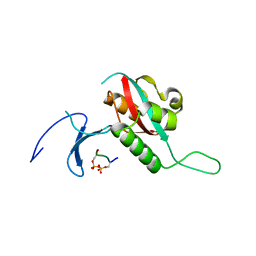 | | STRUCTURAL BASIS FOR THE PHOSPHOSERINE-PROLINE RECOGNITION BY GROUP IV WW DOMAINS | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1, Y(SEP)PT(SEP)S PEPTIDE | | Authors: | Verdecia, M.A, Bowman, M.E, Lu, K.P, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for phosphoserine-proline recognition by group IV WW domains.
Nat.Struct.Biol., 7, 2000
|
|
4R1T
 
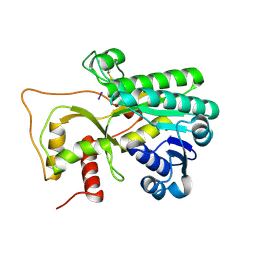 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | cinnamoyl CoA reductase, molecular iodine | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
4R1U
 
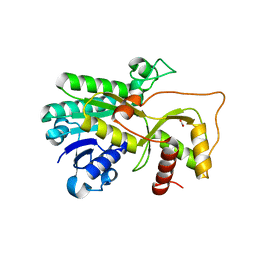 | | Crystal structure of Medicago truncatula cinnamoyl-CoA reductase | | Descriptor: | ACETATE ION, Cinnamoyl CoA reductase | | Authors: | Noel, J.P, Bomati, E.K, Louie, G.V, Bowman, M.E. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
4R1S
 
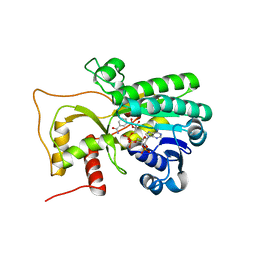 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, cinnamoyl CoA reductase | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
6CJO
 
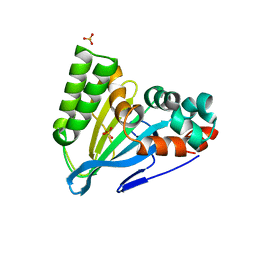 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95S mutation. | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6CJN
 
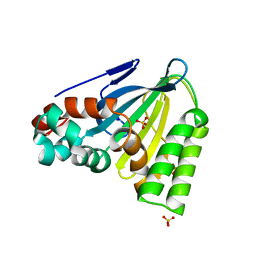 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95T mutation | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
1XDT
 
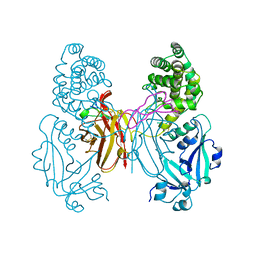 | | COMPLEX OF DIPHTHERIA TOXIN AND HEPARIN-BINDING EPIDERMAL GROWTH FACTOR | | Descriptor: | DIPHTHERIA TOXIN, HEPARIN-BINDING EPIDERMAL GROWTH FACTOR | | Authors: | Louie, G.V, Yang, W, Bowman, M.E, Choe, S. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of diphtheria toxin with an extracellular fragment of its receptor.
Mol.Cell, 1, 1997
|
|
1ZCN
 
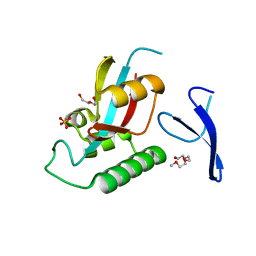 | | human Pin1 Ng mutant | | Descriptor: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-04-12 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1JX1
 
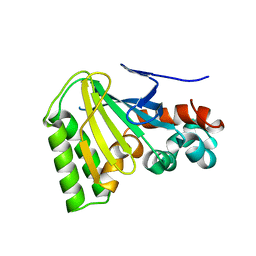 | | Chalcone Isomerase--T48A mutant | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, CHALCONE--FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Bowman, M.E, Noel, J.P. | | Deposit date: | 2001-09-05 | | Release date: | 2002-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Hydrogen Bonds in the Reaction Mechanism of Chalcone Isomerase
Biochemistry, 41, 2002
|
|
1JX0
 
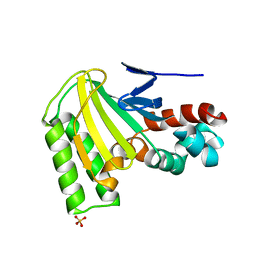 | | Chalcone Isomerase--Y106F mutant | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, CHALCONE--FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Bowman, M.E, Noel, J.P. | | Deposit date: | 2001-09-05 | | Release date: | 2002-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Role of Hydrogen Bonds in the Reaction Mechanism of Chalcone Isomerase
Biochemistry, 41, 2002
|
|
1KNJ
 
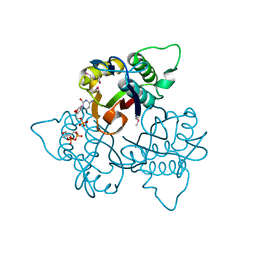 | | Co-Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli Involved in Mevalonate-Independent Isoprenoid Biosynthesis, Complexed with CMP/MECDP/Mn2+ | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
1KNK
 
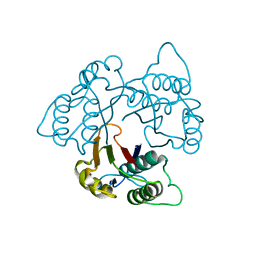 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli involved in Mevalonate-Independent Isoprenoid Biosynthesis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MANGANESE (II) ION | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
4DOL
 
 | |
4DOI
 
 | |
3P9C
 
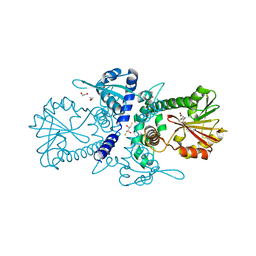 | | Crystal structure of perennial ryegrass LpOMT1 bound to SAH | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
3P9I
 
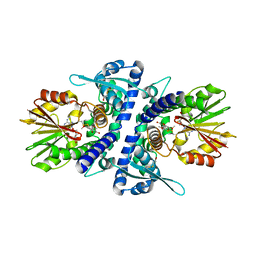 | | Crystal structure of perennial ryegrass LpOMT1 complexed with S-adenosyl-L-homocysteine and sinapaldehyde | | Descriptor: | (2E)-3-(4-hydroxy-3,5-dimethoxyphenyl)prop-2-enal, BETA-MERCAPTOETHANOL, Caffeic acid O-methyltransferase, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
1QLV
 
 | |
1ND7
 
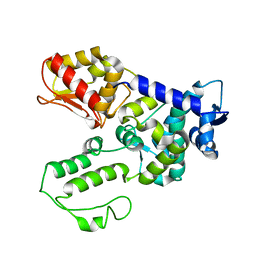 | | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase | | Descriptor: | WW domain-containing protein 1 | | Authors: | Verdecia, M.A, Joaziero, C.A.P, Wells, N.J, Ferrer, J.-L, Bowman, M.E, Hunter, T, Noel, J.P. | | Deposit date: | 2002-12-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase
Mol.Cell, 11, 2003
|
|
1OSH
 
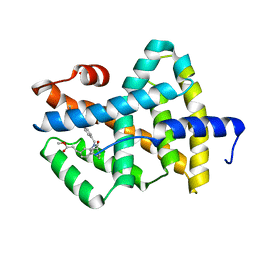 | | A Chemical, Genetic, and Structural Analysis of the nuclear bile acid receptor FXR | | Descriptor: | Bile acid receptor, METHYL 3-{3-[(CYCLOHEXYLCARBONYL){[4'-(DIMETHYLAMINO)BIPHENYL-4-YL]METHYL}AMINO]PHENYL}ACRYLATE | | Authors: | Downes, M, Verdecia, M.A, Roecker, A.J, Hughes, R, Hogenesch, J.B, Kast-Woelbern, H.R, Bowman, M.E, Ferrer, J.-L, Anisfeld, A.M, Edwards, P.A, Rosenfeld, J.M, Alvarez, J.G.A, Noel, J.P, Nicolaou, K.C, Evans, R.M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
Mol.Cell, 11, 2003
|
|
