1ATA
 
 | |
4GU8
 
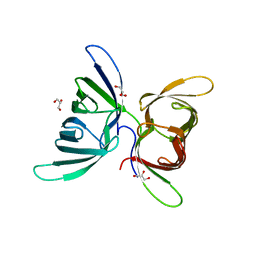 | | Crystal Structure of Burkholderia oklahomensis agglutinin (BOA) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Burkholderia oklahomensis agglutinin (BOA), GLYCEROL | | Authors: | Whitley, M.J, Furey, W, Gronenborn, A.M. | | Deposit date: | 2012-08-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Burkholderia oklahomensis agglutinin is a canonical two-domain OAA-family lectin: structures, carbohydrate binding and anti-HIV activity.
Febs J., 280, 2013
|
|
2EZN
 
 | |
2L9Y
 
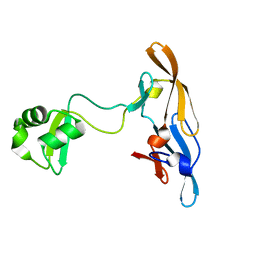 | | Solution structure of the MoCVNH-LysM module from the rice blast fungus Magnaporthe oryzae protein (MGG_03307) | | Descriptor: | CVNH-LysM lectin | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Montanini, B, Kershaw, M.J, Talbot, N.J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2011-02-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-Function Analysis of a CVNH-LysM Lectin Expressed during Plant Infection by the Rice Blast Fungus Magnaporthe oryzae.
Structure, 19, 2011
|
|
1BDS
 
 | |
4EMW
 
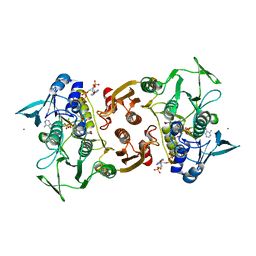 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor EtVC-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Edwards, J.S, Wallace, B.D, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-12 | | Release date: | 2012-10-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4GAT
 
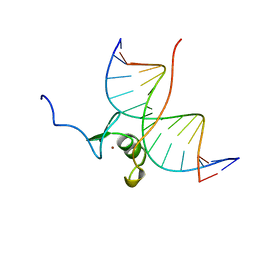 | | SOLUTION NMR STRUCTURE OF THE WILD TYPE DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13BP DNA CONTAINING A CGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*GP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors.
J.Mol.Biol., 277, 1998
|
|
4GR7
 
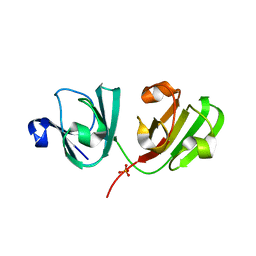 | | The human W42R Gamma D-Crystallin Mutant Structure at 1.7A Resolution | | Descriptor: | Gamma-crystallin D, PHOSPHATE ION | | Authors: | Ji, F, Jung, J, Koharudin, L.M.I, Gronenborn, A.M. | | Deposit date: | 2012-08-24 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The human W42R gamma D-crystallin mutant structure provides a link between congenital and age-related cataracts.
J.Biol.Chem., 288, 2013
|
|
1MDI
 
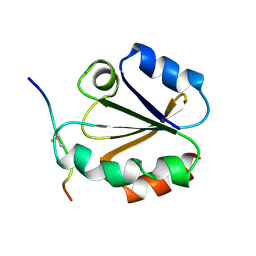 | |
1MDK
 
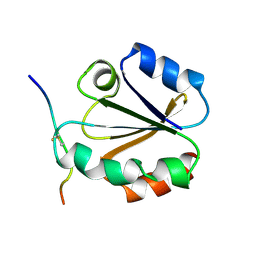 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN NFKB (RESIDUES 56-68 OF THE P50 SUBUNIT OF NFKB) | | Descriptor: | TARGET SITE IN HUMAN NFKB, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1995-02-27 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B.
Structure, 3, 1995
|
|
1MDJ
 
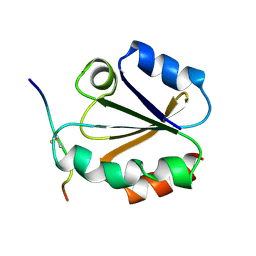 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN NFKB (RESIDUES 56-68 OF THE P50 SUBUNIT OF NFKB) | | Descriptor: | TARGET SITE IN HUMAN NFKB, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1995-02-27 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B.
Structure, 3, 1995
|
|
2NEF
 
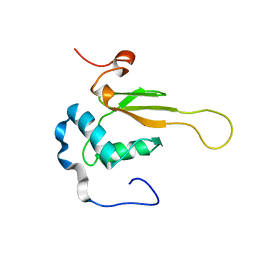 | | HIV-1 NEF (REGULATORY FACTOR), NMR, 40 STRUCTURES | | Descriptor: | NEGATIVE FACTOR (F-PROTEIN) | | Authors: | Grzesiek, S, Bax, A, Clore, G.M, Gronenborn, A.M, Hu, J.S, Kaufman, J, Palmer, I, Stahl, S.J, Tjandra, N, Wingfield, P.T. | | Deposit date: | 1997-02-12 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and backbone dynamics of HIV-1 Nef.
Protein Sci., 6, 1997
|
|
1CBH
 
 | |
1YUI
 
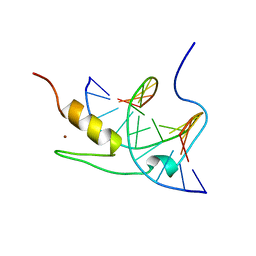 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1996-12-31 | | Release date: | 1997-12-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
1YUJ
 
 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, 50 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1996-12-31 | | Release date: | 1997-12-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
1G6E
 
 | | ANTIFUNGAL PROTEIN FROM STREPTOMYCES TENDAE TU901, 30-CONFORMERS ENSEMBLE | | Descriptor: | ANTIFUNGAL PROTEIN | | Authors: | Campos-Olivas, R, Bormann, C, Hoerr, I, Jung, G, Gronenborn, A.M. | | Deposit date: | 2000-11-04 | | Release date: | 2001-03-28 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics and chitin binding of the anti-fungal protein from Streptomyces tendae TU901.
J.Mol.Biol., 308, 2001
|
|
4EQX
 
 | | Crystal Structure of the C43S Mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Edwards, J.S, Wallace, B.D, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
1GH5
 
 | | ANTIFUNGAL PROTEIN FROM STREPTOMYCES TENDAE TU901, NMR AVERAGE STRUCTURE | | Descriptor: | ANTIFUNGAL PROTEIN | | Authors: | Campos-Olivas, R, Bormann, C, Hoerr, I, Jung, G, Gronenborn, A.M. | | Deposit date: | 2000-11-04 | | Release date: | 2001-03-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics and chitin binding of the anti-fungal protein from Streptomyces tendae TU901.
J.Mol.Biol., 308, 2001
|
|
1NCP
 
 | |
1N02
 
 | |
1OLG
 
 | |
4EQW
 
 | | Crystal Structure of the Y361F, Y419F Mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, COENZYME A, Coenzyme A disulfide reductase, ... | | Authors: | Edwards, J.S, Wallace, B.D, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4EM4
 
 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor Pethyl-VS-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wallace, B.D, Edwards, J.S, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4EM3
 
 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor MeVS-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wallace, B.D, Edwards, J.S, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4EQS
 
 | | Crystal structure of the Y419F mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, COENZYME A, Coenzyme A disulfide reductase, ... | | Authors: | Wallace, B.D, Edwards, J.S, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
