8DAE
 
 | |
1HH1
 
 | | THE STRUCTURE OF HJC, A HOLLIDAY JUNCTION RESOLVING ENZYME FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | HOLLIDAY JUNCTION RESOLVING ENZYME HJC | | Authors: | Bond, C.S, Kvaratskhelia, M, Richard, D, White, M.F, Hunter, W.N. | | Deposit date: | 2000-12-18 | | Release date: | 2001-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Hjc, a Holliday Junction Resolvase, from Sulfolobus Solfataricus
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GXF
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN COMPLEX WITH THE INHIBITOR QUINACRINE MUSTARD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALEIC ACID, QUINACRINE MUSTARD, ... | | Authors: | Bond, C.S, Peterson, M.R, Vickers, T.J, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 2002-04-04 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two Interacting Binding Sites for Quinacrine Derivatives in the Active Site of Trypanothione Reductase: A Template for Drug Design
J.Biol.Chem., 279, 2004
|
|
4TKD
 
 | | Sulfolobus solfataricus HJC mutants | | Descriptor: | Holliday junction resolvase Hjc | | Authors: | Bond, C.S. | | Deposit date: | 2014-05-26 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Unengineering: Reducing the Crystallisability of Sulfolobus solfataricus Hjc
Aust.J.Chem., 67, 2014
|
|
4TKK
 
 | | Sulfolobus solfataricus HJC mutants | | Descriptor: | Holliday junction resolvase Hjc | | Authors: | Bond, C.S. | | Deposit date: | 2014-05-27 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Unengineering: Reducing the Crystallisability of Sulfolobus solfataricus Hjc
Aust.J.Chem., 67, 2014
|
|
6DHI
 
 | |
6AZT
 
 | |
1E9W
 
 | | Structure of the macrocycle thiostrepton solved using the anomalous dispersive contribution from sulfur | | Descriptor: | THIOSTREPTON | | Authors: | Bond, C.S, Shaw, M.P, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2000-10-27 | | Release date: | 2001-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the Macrocycle Thiostrepton Solved Using the Anomalous Dispersive Contribution from Sulfur
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1E58
 
 | | E.coli cofactor-dependent phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCERATE MUTASE, SULFATE ION | | Authors: | Bond, C.S, Hunter, W.N. | | Deposit date: | 2000-07-19 | | Release date: | 2001-03-20 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Structure of the Phosphohistidine-Activated Form of Escherichia Coli Cofactor-Dependent Phosphoglycerate Mutase.
J.Biol.Chem., 276, 2001
|
|
1E59
 
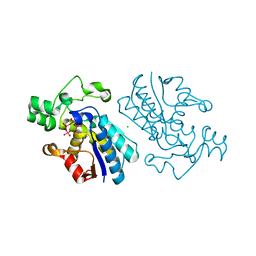 | |
1BAW
 
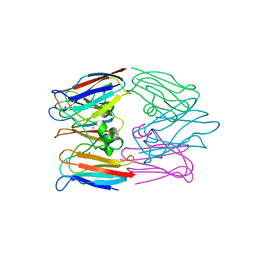 | | PLASTOCYANIN FROM PHORMIDIUM LAMINOSUM | | Descriptor: | COPPER (II) ION, PLASTOCYANIN, ZINC ION | | Authors: | Bond, C.S, Guss, J.M, Freeman, H.C, Wagner, M.J, Howe, C.J, Bendall, D.S. | | Deposit date: | 1998-04-19 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of plastocyanin from the cyanobacterium Phormidium laminosum.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BZL
 
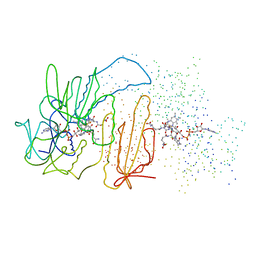 | | CRYSTAL STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN COMPLEX WITH TRYPANOTHIONE, AND THE STRUCTURE-BASED DISCOVERY OF NEW NATURAL PRODUCT INHIBITORS | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE REDUCTASE (OXIDIZED FORM) | | Authors: | Bond, C.S, Zhang, Y, Berriman, M, Cunningham, M, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 1998-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Trypanosoma cruzi trypanothione reductase in complex with trypanothione, and the structure-based discovery of new natural product inhibitors.
Structure Fold.Des., 7, 1999
|
|
1AOG
 
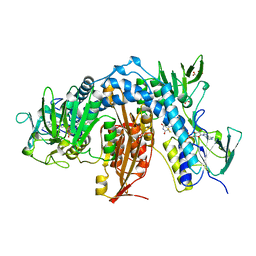 | |
1QHQ
 
 | | AURACYANIN, A BLUE COPPER PROTEIN FROM THE GREEN THERMOPHILIC PHOTOSYNTHETIC BACTERIUM CHLOROFLEXUS AURANTIACUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (AURACYANIN), ... | | Authors: | Bond, C.S, Blankenship, R.E, Freeman, H.C, Guss, J.M, Maher, M, Selvaraj, F, Wilce, M.C.J, Willingham, K. | | Deposit date: | 1999-05-25 | | Release date: | 2001-03-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of auracyanin, a "blue" copper protein from the green thermophilic photosynthetic bacterium Chloroflexus aurantiacus.
J.Mol.Biol., 306, 2001
|
|
7MMY
 
 | |
8ECG
 
 | | Complex of HMG1 with pitavastatin | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1, GLYCEROL, Pitavastatin | | Authors: | Haywood, J, Bond, C.S. | | Deposit date: | 2022-09-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.
Nat Commun, 13, 2022
|
|
1GY8
 
 | | Trypanosoma brucei UDP-galactose 4' epimerase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE 4-EPIMERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Shaw, M.P, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-04-21 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Resolution Crystal Structure of Trypanosoma Brucei Udp-Galactose 4'-Epimerase: A Potential Target for Structure-Based Development of Novel Trypanocides
Mol.Biochem.Parasitol., 126, 2003
|
|
6EEN
 
 | |
6WCK
 
 | | KRAS G-quadruplex G16T mutant with Bromo Uracil replacing T8 and T16. | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*CP*GP*GP*(BRU)P*GP*TP*GP*GP*GP*AP*AP*(BRU)P*AP*GP*GP*GP*AP*A)-3'), POTASSIUM ION | | Authors: | Schmidberger, J.W, Ou, A, Smith, N.M, Iyer, K.S, Bond, C.S. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition.
Nucleic Acids Res., 48, 2020
|
|
4MTT
 
 | | Ni- and Zn-bound GloA2 at low resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Lactoylglutathione lyase, ... | | Authors: | Bythell-Douglas, R, Bond, C.S. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The crystal structure of a homodimeric Pseudomonas glyoxalase I enzyme reveals asymmetric metallation commensurate with half-of-sites activity.
Chemistry, 21, 2015
|
|
6XT6
 
 | | pro-concanavalin A: Precursor of circularly permuted concanavalin A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Concanavalin-A, ... | | Authors: | Nonis, S.G, Haywood, J, Schmidberger, J.W, Bond, C.S. | | Deposit date: | 2020-07-17 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural and biochemical analyses of concanavalin A circular permutation by jack bean asparaginyl endopeptidase.
Plant Cell, 33, 2021
|
|
6XT5
 
 | |
6WMZ
 
 | | Crystal structure of human SFPQ/NONO complex | | Descriptor: | Non-POU domain-containing octamer-binding protein, SULFATE ION, Splicing factor, ... | | Authors: | Lee, M, Bond, C.S. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure analysis of human SFPQ/NONO complex
To Be Published
|
|
8F2G
 
 | | Crystal structure of Hen Egg White Lysozyme at 0.44 GPa | | Descriptor: | Lysozyme C | | Authors: | Marshall, A.C, Boer, S.A, Turner, G, Moggach, S.A, Bond, C.S, Vrielink, A. | | Deposit date: | 2022-11-08 | | Release date: | 2022-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | High-pressure single-crystal diffraction at the Australian Synchrotron.
J.Synchrotron Radiat., 30, 2023
|
|
7MPY
 
 | |
