7O2L
 
 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
6QSP
 
 | | Ketosynthase (ApeO) in Complex with its Chain Length Factor (ApeC) from Xenorhabdus doucetiae | | Descriptor: | Beta-ketoacyl synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grammbitter, G.L.C, Schmalhofer, M, Groll, M, Bode, H. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments.
J.Am.Chem.Soc., 141, 2019
|
|
6EWS
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | Descriptor: | NRPS Kj12A-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6QSR
 
 | | The Dehydratase Heterocomplex ApeI:P from Xenorhabdus doucetiae | | Descriptor: | AMP-dependent synthetase and ligase, Beta-hydroxyacyl-(Acyl-carrier-protein) dehydratase FabA/FabZ, SODIUM ION | | Authors: | Grammbitter, G.L.C, Schmalhofer, M, Groll, M, Bode, H. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments.
J.Am.Chem.Soc., 141, 2019
|
|
6HXA
 
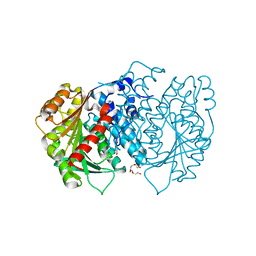 | | AntI from P. luminescens catalyses terminal polyketide shortening in the biosynthesis of anthraquinones | | Descriptor: | AntI, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Zhou, Q, Braeuer, A, Grammbitter, G, Schmalhofer, M, Saura, P, Adihou, H, Kaila, V.R.I, Groll, M, Bode, H. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of polyketide shortening in anthraquinone biosynthesis ofPhotorhabdus luminescens.
Chem Sci, 10, 2019
|
|
8BW1
 
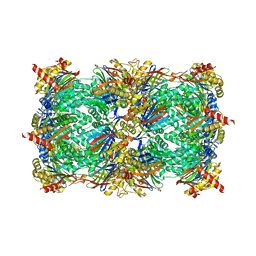 | | Yeast 20S proteasome in complex with an engineered fellutamide derivative (C14QAL) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bozhueyuek, K.A.J, Praeve, L, Kegler, C, Kaiser, S, Shi, Y, Kuttenlochner, W, Schenk, L, Groll, M, Hochberg, G.K.A, Bode, H.B. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evolution-inspired engineering of nonribosomal peptide synthetases.
Science, 383, 2024
|
|
6TRP
 
 | | Solution Structure of Docking Domain Complex of Pax NRPS: PaxC NDD - PaxB CDD | | Descriptor: | Peptide synthetase XpsB,Peptide synthetase XpsB | | Authors: | Watzel, J, Hacker, C, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A New Docking Domain Type in the Peptide-Antimicrobial-Xenorhabdus Peptide Producing Nonribosomal Peptide Synthetase fromXenorhabdus bovienii.
Acs Chem.Biol., 15, 2020
|
|
6EWT
 
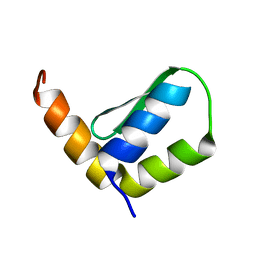 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | Descriptor: | NRPS Kj12B-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWU
 
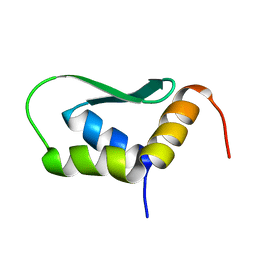 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12C-NDD | | Descriptor: | NRPS Kj12C-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWV
 
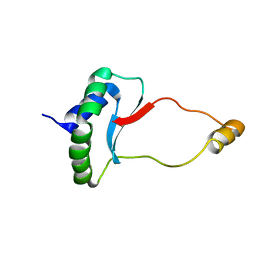 | | Solution Structure of Docking Domain Complex of RXP NRPS: Kj12C NDD - Kj12B CDD | | Descriptor: | NRPS Kj12C-NDD, NRPS Kj12B-CDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6SMP
 
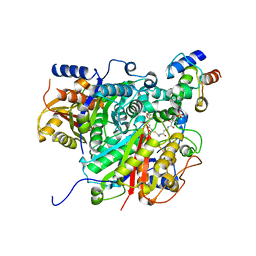 | | AntDE:AntF (holo): type II PKS acyl-carrier protein in complex with its ketosynthase bound to the hexaketide | | Descriptor: | (2~{R})-3,3-dimethyl-2-oxidanyl-~{N}-[3-oxidanylidene-3-[2-[(1~{R},4~{Z},6~{Z},8~{Z})-1,5,7,9-tetrakis(oxidanyl)-3,11-bis(oxidanylidene)dodeca-4,6,8-trienyl]sulfanylethylamino]propyl]-4-[tris(oxidanyl)-$l^{5}-phosphanyl]oxy-butanamide, Acyl carrier protein, Ketoacyl_synth_N domain-containing protein, ... | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
6SMO
 
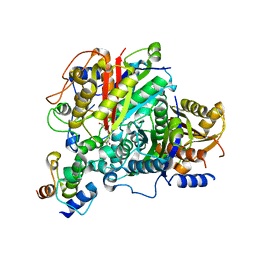 | | AntDE:AntF (apo): type II PKS acyl-carrier protein in complex with its ketosynthase bound to the hexaketide | | Descriptor: | 3,5,7,9,11-pentakis(oxidanylidene)dodecanal, Acyl carrier protein, Ketoacyl_synth_N domain-containing protein, ... | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
6SM4
 
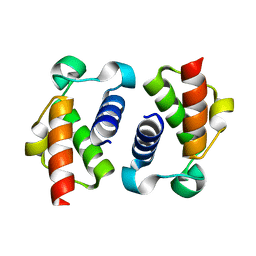 | | AntF (apo): type II PKS acyl-carrier protein | | Descriptor: | Acyl carrier protein | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
6SMD
 
 | | PlMCAT:AntF (holo): type II PKS acyl-carrier protein in complex with its malonyl-transacylase | | Descriptor: | Acyl carrier protein, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
6SM6
 
 | | AntF (holo): type II PKS acyl-carrier protein | | Descriptor: | Acyl carrier protein | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
8QD8
 
 | | Wdyg1p in complex with 1,3-Dihydroxynaphthalene | | Descriptor: | 1,2-ETHANEDIOL, Yellowish-green 1-like protein, naphthalene-1,3-diol | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD2
 
 | | Ayg1p in complex with 1,3-Dihydroxynaphthalene | | Descriptor: | Pigment biosynthesis protein yellowish-green 1, naphthalene-1,3-diol | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD1
 
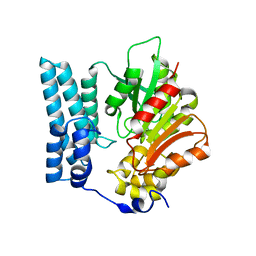 | | Ayg1p from A. fumigatus catalyzes polyketide shortening in the biosynthesis of DHN-melanin | | Descriptor: | Pigment biosynthesis protein yellowish-green 1 | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD4
 
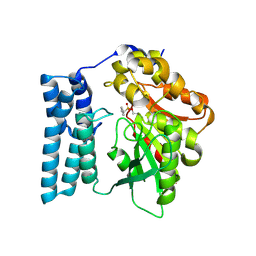 | | Ayg1p active site converted to tetrahedral sulfonate ester | | Descriptor: | Pigment biosynthesis protein yellowish-green 1, phenylmethanesulfonic acid | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD3
 
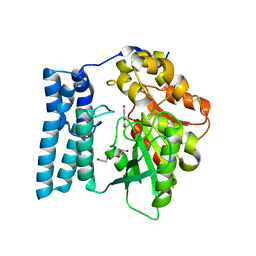 | | Ayg1p in complex with 1,3,6,8-Tetrahydroxynaphthalene | | Descriptor: | 1,2-ETHANEDIOL, FLAVIOLIN, Pigment biosynthesis protein yellowish-green 1, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD9
 
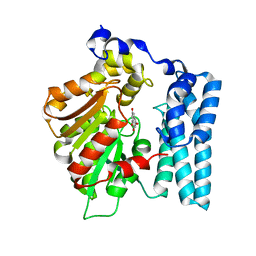 | | Wdyg1p in complex with 1,3,6,8-Tetrahydroxynaphthalene | | Descriptor: | 1,2-ETHANEDIOL, Yellowish-green 1-like protein, naphthalene-1,3,6,8-tetrol | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QDA
 
 | | Wdyg1p Ser256DHA (PMSF) | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, Yellowish-green 1-like protein | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
6ZP8
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB335 | | Descriptor: | (2~{S},3~{R})-~{N}-[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]-3-oxidanyl-2-(3-phenylpropanoylamino)butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
6ZOU
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB333 | | Descriptor: | 11-methyl-~{N}-[(2~{S},3~{R})-1-[[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]amino]-3-oxidanyl-1-oxidanylidene-butan-2-yl]dodecanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
6ZP6
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB334 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
