7B3K
 
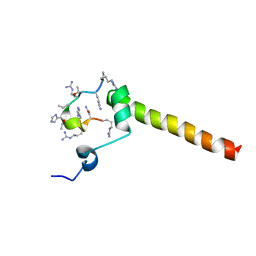 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
7B3J
 
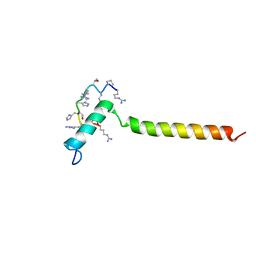 | | Dynamic complex between all-D-enantiomeric peptide D3 with wild-type amyloid precursor protein 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-12-08 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
1RQT
 
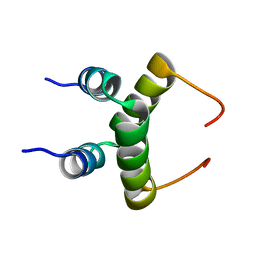 | | NMR structure of dimeric N-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQU
 
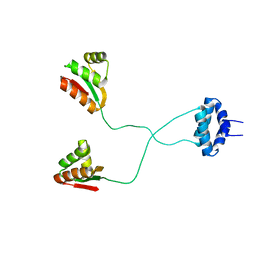 | | NMR structure of L7 dimer from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQV
 
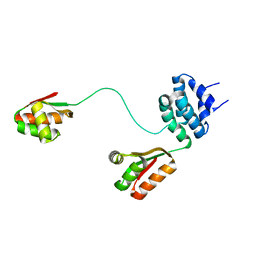 | | Spatial model of L7 dimer from E.coli with one hinge region in helical state | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome
J.Biol.Chem., 279, 2004
|
|
1RQS
 
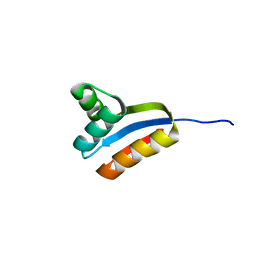 | | NMR structure of C-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
2J5D
 
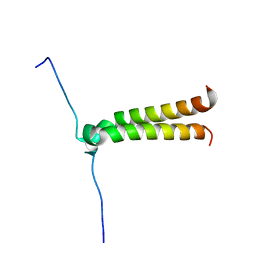 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | Descriptor: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | Authors: | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | Deposit date: | 2006-09-14 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
7O9U
 
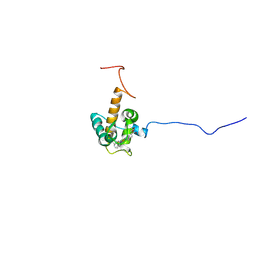 | | Solution structure of oxidized cytochrome c552 from Thioalkalivibrio paradoxus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Britikov, V.V, Britikova, E.V, Altukhov, D.A, Timofeev, V.I, Dergousova, N.I, Rakitina, T.V, Tikhonova, T.V, Usanov, S.A, Popov, V.O, Bocharov, E.V. | | Deposit date: | 2021-04-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Unusual Cytochrome c 552 from Thioalkalivibrio paradoxus : Solution NMR Structure and Interaction with Thiocyanate Dehydrogenase.
Int J Mol Sci, 23, 2022
|
|
1J5J
 
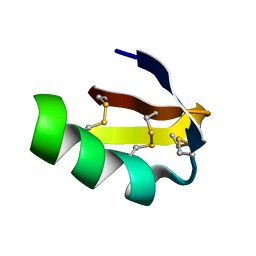 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
6YTS
 
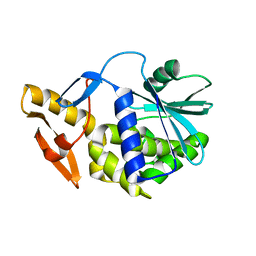 | | Solution NMR structure of type-I ribosome-inactivating protein trichobakin (TBK) | | Descriptor: | Trichobakin | | Authors: | Britikov, V.V, Bocharov, E.V, Britikova, E.V, Le, T.B.T, Phan, C.V, Boyko, K.M, Arseniev, A.S, Usanov, S.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of type-I ribosome-inactivating protein trichobakin (TBK)
To Be Published
|
|
6Z1Y
 
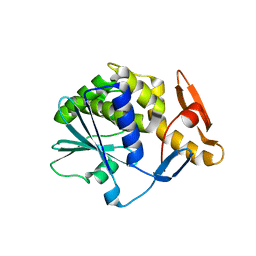 | | Crystal structure of type-I ribosome-inactivating protein trichobakin (TBK) | | Descriptor: | SODIUM ION, Trichobakin | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Britikov, V.V, Bocharov, E.V, Britikova, E.V, Le, T.B.T, Phan, C.V, Popov, V.O, Usanov, S.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of type-I ribosome-inactivating protein trichobakin (TBK)
To Be Published
|
|
1LGL
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
1FFJ
 
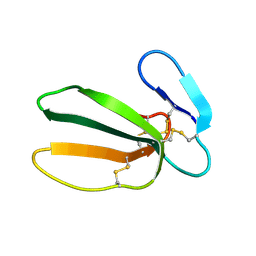 | | NMR STRUCTURE OF CARDIOTOXIN IN DPC-MICELLE | | Descriptor: | CYTOTOXIN 2 | | Authors: | Dubovskii, P.V, Dementieva, D.V, Bocharov, E.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2000-07-25 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Membrane binding motif of the P-type cardiotoxin.
J.Mol.Biol., 305, 2001
|
|
5OHD
 
 | | Putative inactive (dormant) dimeric state of GHR transmembrane domain | | Descriptor: | Growth hormone receptor | | Authors: | Lesovoy, D.M, Bocharov, E.V, Bocharova, O.V, Urban, A.S, Arseniev, A.S. | | Deposit date: | 2017-07-15 | | Release date: | 2018-04-11 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the signal transduction via transmembrane domain of the human growth hormone receptor.
Biochim. Biophys. Acta, 1862, 2018
|
|
5OEK
 
 | | Putative active dimeric state of GHR transmembrane domain | | Descriptor: | Growth hormone receptor | | Authors: | Lesovoy, D.M, Bocharov, E.V, Bocharova, O.V, Urban, A.S, Arseniev, A.S. | | Deposit date: | 2017-07-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the signal transduction via transmembrane domain of the human growth hormone receptor.
Biochim. Biophys. Acta, 1862, 2018
|
|
2LLM
 
 | |
7PH8
 
 | |
7PL4
 
 | |
7PHT
 
 | |
6ZYG
 
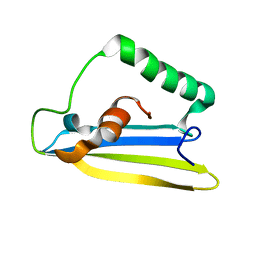 | | Emfourin (M4in) from Serratia proteamaculans, M4 family peptidase inhibitor | | Descriptor: | Protealysin-associated protein | | Authors: | Bozin, T.N, Chukhontseva, K.N, Konarev, P.V, Bocharov, E.V, Demidyuk, I.V. | | Deposit date: | 2020-07-31 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | NMR structure of emfourin, a novel metalloprotease inhibitor
To Be Published
|
|
9KK0
 
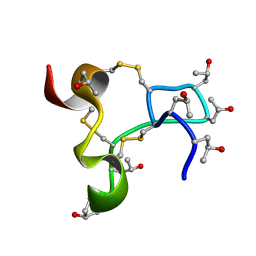 | | Solution structure of kappa-conotoxin RIIIJ | | Descriptor: | Kappa-conotoxin RIIIJ | | Authors: | Lushpa, V.A, Vassilevski, A.A, Bocharov, E.V, Olivera, B.M, Rivier, J.E, Raghuraman, S. | | Deposit date: | 2024-11-12 | | Release date: | 2024-12-18 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kappa-conotoxin RIIIJ
To Be Published
|
|
9KK3
 
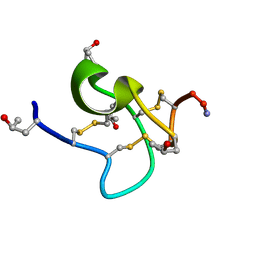 | | Solution structure of kappa-conotoxin RIIIK | | Descriptor: | Kappa-conotoxin RIIIK | | Authors: | Lushpa, V.A, Vassilevski, A.A, Bocharov, E.V, Olivera, B.M, Rivier, J, Raghuraman, S. | | Deposit date: | 2024-11-12 | | Release date: | 2024-12-18 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kappa-conotoxin RIIIK
To Be Published
|
|
2LCX
 
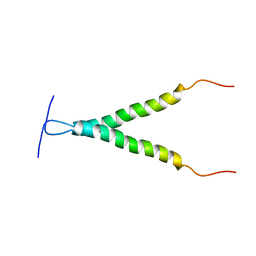 | |
2L2T
 
 | |
1CCQ
 
 | |
