4MHC
 
 | | Crystal Structure of a Nucleoporin | | Descriptor: | Nucleoporin NUP157 | | Authors: | Seo, H.S, Blus, B.J, Blobel, G. | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and nucleic acid binding activity of the nucleoporin Nup157.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4WNL
 
 | | The X-ray structure of a RNA-binding protein complex | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Singh, N, Blobel, G, Shi, H. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hooking She3p onto She2p for myosin-mediated cytoplasmic mRNA transport.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QUV
 
 | | Structure of an integral membrane delta(14)-sterol reductase | | Descriptor: | Delta(14)-sterol reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, X, Blobel, G. | | Deposit date: | 2014-07-12 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum.
Nature, 517, 2015
|
|
2XU7
 
 | | Structural basis for RbAp48 binding to FOG-1 | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, TETRAETHYLENE GLYCOL, ZINC FINGER PROTEIN ZFPM1 | | Authors: | Lejon, S, Thong, S.Y, Murthy, A, Blobel, G.A, Mackay, J.P, Murzina, N.V, Laue, E.D. | | Deposit date: | 2010-10-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights Into Association of the Nurd Complex with Fog-1 from the Crystal Structure of an Rbap48-Fog- 1 Complex.
J.Biol.Chem., 286, 2011
|
|
1QBK
 
 | |
1XKS
 
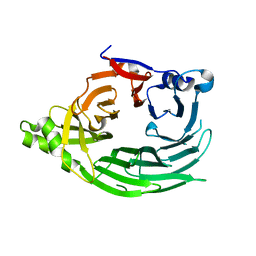 | |
6BGG
 
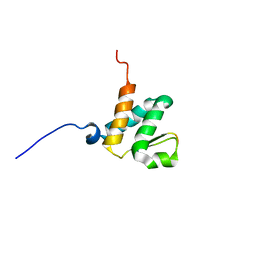 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
7RDN
 
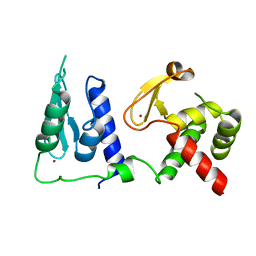 | | Crystal structure of S. cerevisiae pre-mRNA leakage protein 39 (Pml39) | | Descriptor: | Pre-mRNA leakage protein 39, ZINC ION | | Authors: | Hashimoto, H, Ramirez, D.H, Pawlak, N, Blobel, G, Palancade, B, Debler, E.W. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules.
Sci Rep, 12, 2022
|
|
2H94
 
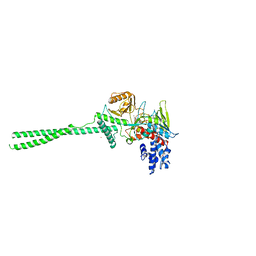 | |
3OPB
 
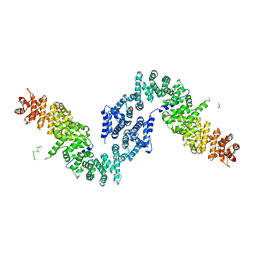 | | Crystal structure of She4p | | Descriptor: | SWI5-dependent HO expression protein 4 | | Authors: | Shi, H, Blobel, G. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
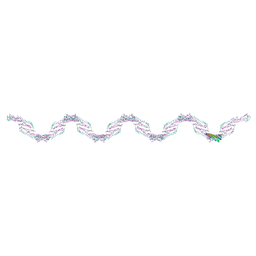
3T98
 
 | |
3T97
 
 | |
4OWR
 
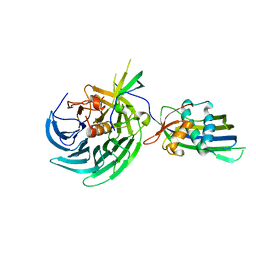 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | Descriptor: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | Authors: | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | Deposit date: | 2014-02-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1BK6
 
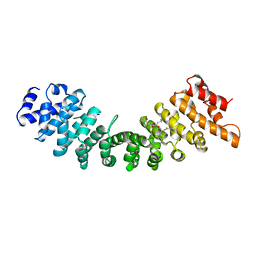 | | KARYOPHERIN ALPHA (YEAST) + SV40 T ANTIGEN NLS | | Descriptor: | KARYOPHERIN ALPHA, LARGE T ANTIGEN | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BK5
 
 | | KARYOPHERIN ALPHA FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | COBALT (II) ION, KARYOPHERIN ALPHA | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
4LL7
 
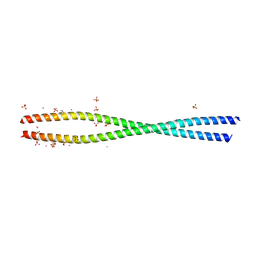 | | Structure of She3p amino terminus. | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DYSPROSIUM ION, ... | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3F3P
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21212 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3F3F
 
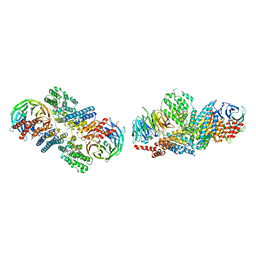 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
4J3H
 
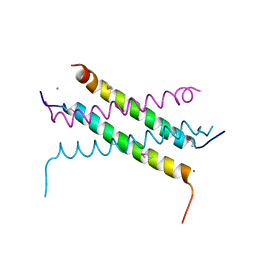 | |
2MPL
 
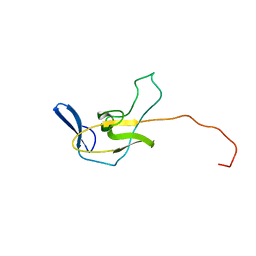 | |
2OSZ
 
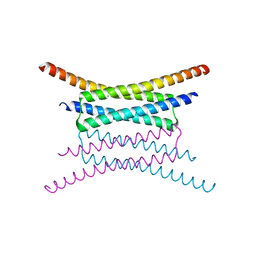 | |
5SUP
 
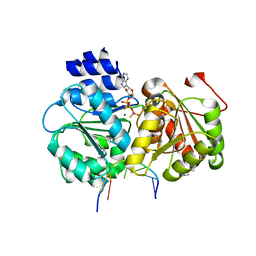 | | Crystal structure of the Sub2-Yra1 complex in association with RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase SUB2, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ren, Y, Schmiege, P, Blobel, G. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
5U73
 
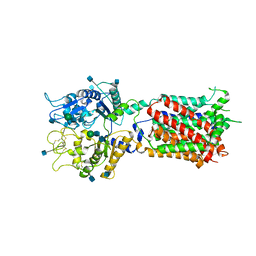 | | Crystal structure of human Niemann-Pick C1 protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Niemann-Pick C1 protein, ... | | Authors: | Li, X, Wang, J, Blobel, G. | | Deposit date: | 2016-12-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4GQ2
 
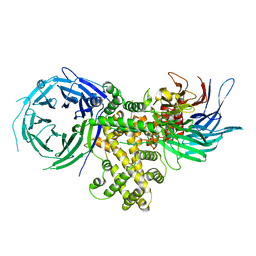 | | S. pombe Nup120-Nup37 complex | | Descriptor: | Nucleoporin nup120, Nup37 | | Authors: | Liu, X, Mitchell, J, Wozniak, R, Blobel, G, Fan, J. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evolution of the membrane-coating module of the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2OIT
 
 | |
