4ZFK
 
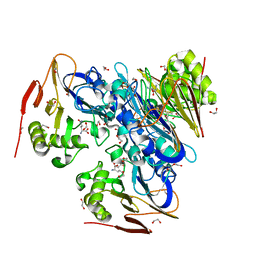 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC with glutamine | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase EgtC, GLUTAMINE | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
4ZFJ
 
 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC, apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Amidohydrolase EgtC | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
4ZFL
 
 | |
9F92
 
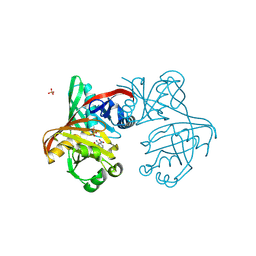 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-nitrobenzoic acid | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-azanyl-3-nitro-benzoic acid, SULFATE ION, ... | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F96
 
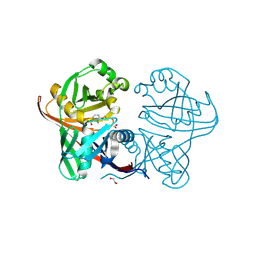 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-ethoxybenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-3-ethoxy-benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F94
 
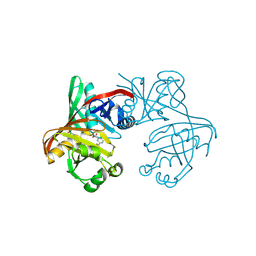 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-5-(3-hydroxyphenyl)benzoic acid | | Descriptor: | 2-azanyl-5-(3-hydroxyphenyl)benzoic acid, ACETATE ION, FORMIC ACID, ... | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F95
 
 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-hydroxy-5-(3-hydroxyphenyl)benzoic acid | | Descriptor: | 2-azanyl-5-(3-hydroxyphenyl)-3-oxidanyl-benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F93
 
 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-5-(4-fluorophenyl)benzoic acid | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-azanyl-5-(4-fluorophenyl)benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
6YHL
 
 | |
6YHN
 
 | | Crystal structure of domains 4-5 of CNFy from Yersinia pseudotuberculosis | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, Cytotoxic necrotizing factor, ... | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
6YHM
 
 | | Crystal structure of the C-terminal domain of CNFy from Yersinia pseudotuberculosis | | Descriptor: | Cytotoxic necrotizing factor, MAGNESIUM ION | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
8AJQ
 
 | |
6YHK
 
 | | Crystal structure of full-length CNFy (C866S) from Yersinia pseudotuberculosis | | Descriptor: | CHLORIDE ION, Cytotoxic necrotizing factor, SULFATE ION | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
8AID
 
 | |
7ZPN
 
 | | Crystal Structure of IscR from Dinoroseobacter shibae | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator, SULFATE ION | | Authors: | Lukat, P, Ploetzky, L, Blankenfeldt, W, Jahn, D, Haertig, E. | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dinoroseobacter shibae IscR homolog acts as a repressor for iron acquisition genes
To Be Published
|
|
6EV1
 
 | | Crystal structure of antibody against schizophyllan | | Descriptor: | Heavy chain, Light chain | | Authors: | Sung, K.H, Josewski, J, Dubel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
6ET1
 
 | |
6EYS
 
 | |
6ET2
 
 | |
6EYV
 
 | |
6ET0
 
 | | Crystal structure of PqsBC (C129A) mutant from Pseudomonas aeruginosa (crystal form 1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Witzgall, F, Blankenfeldt, W. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 19, 2018
|
|
6ESZ
 
 | |
6EV2
 
 | | Crystal structure of antibody against schizophyllan in complex with laminarihexaose | | Descriptor: | Heavy chain, Light chain, beta-D-glucopyranose, ... | | Authors: | Sung, K.H, Josewski, J, Duebel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
6ET3
 
 | | Crystal structure of PqsBC (C129S) mutant from Pseudomonas aeruginosa (crystal form 4) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, PqsB, PqsC, ... | | Authors: | Witzgall, F, Blankenfeldt, W. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 19, 2018
|
|
3N6O
 
 | | Crystal structure of the GEF and P4M domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | SULFATE ION, guanine nucleotide exchange factor | | Authors: | Schoebel, S, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA.
Embo Rep., 11, 2010
|
|
