5JGJ
 
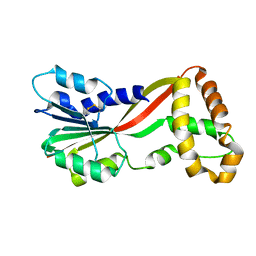 | | Crystal structure of GtmA | | Descriptor: | UbiE/COQ5 family methyltransferase, putative | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
3DSX
 
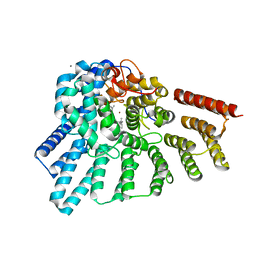 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with di-prenylated peptide Ser-Cys(GG)-Ser-Cys(GG) derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
5HIS
 
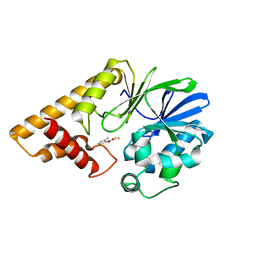 | |
3DH8
 
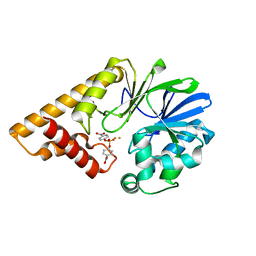 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | FE (III) ION, Uncharacterized protein PA1000, bis(4-nitrophenyl) hydrogen phosphate | | Authors: | Yu, S, Blankenfeldt, W. | | Deposit date: | 2008-06-17 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
3DSW
 
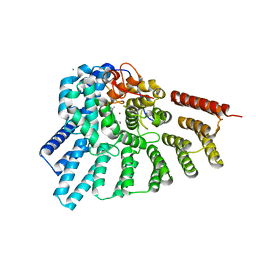 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with mono-prenylated peptide Ser-Cys(GG)-Ser-Cys derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
5K1S
 
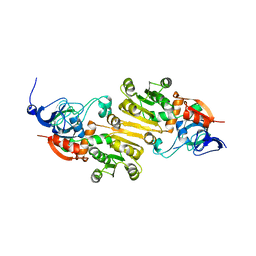 | | crystal structure of AibC | | Descriptor: | Oxidoreductase, zinc-binding dehydrogenase family, ZINC ION | | Authors: | Bock, T, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of AibC, a reductase involved in alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5K7F
 
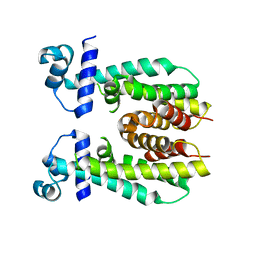 | | Crystal structure of apo AibR | | Descriptor: | ACETATE ION, Transcriptional regulator, TetR family | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
5K7Z
 
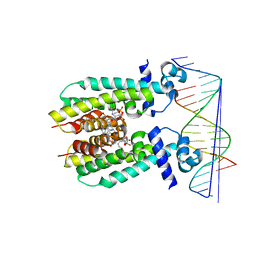 | | Crystal structure of AibR in complex with isovaleryl coenzyme A and operator DNA | | Descriptor: | DNA (32-MER), Isovaleryl-coenzyme A, Transcriptional regulator, ... | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-27 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of AibR in complex with isovaleryl coenzyme A and operator DNA
to be published
|
|
6TAH
 
 | |
6TH5
 
 | |
6TJL
 
 | |
6TSI
 
 | |
6TV9
 
 | |
6TUM
 
 | |
6TPO
 
 | | Conformation of cd1 nitrite reductase NirS without bound heme d1 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, HEME C, Nitrite reductase, ... | | Authors: | Kluenemann, T, Blankenfeldt, W. | | Deposit date: | 2019-12-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Structure of heme d 1 -free cd 1 nitrite reductase NirS.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TUK
 
 | | Crystal structure of Fdr9 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification and crystal structure determination of a ferredoxin reductase from the actinobacterium Thermobifida fusca.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6THO
 
 | |
6TO2
 
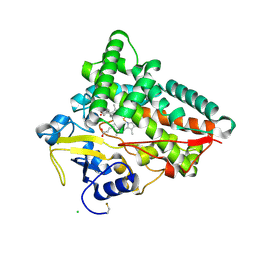 | | Crystal structure of CYP154C5 from Nocardia farcinica in complex with 5alpha-Androstan-3-one | | Descriptor: | (5~{S},8~{S},9~{S},10~{S},13~{S},14~{S})-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydrocyclopenta[a]phenanthren-3-one, CHLORIDE ION, Cytochrome P450 monooxygenase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CYP154C5 Regioselectivity in Steroid Hydroxylation Explored by Substrate Modifications and Protein Engineering*.
Chembiochem, 22, 2021
|
|
6TP9
 
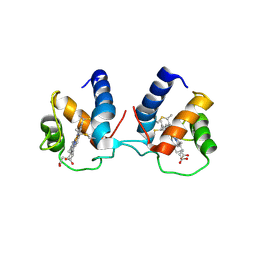 | | c-type cytochrome NirC | | Descriptor: | Cytochrome c55X, HEME C | | Authors: | Kluenemann, T, Henke, S, Blankenfeldt, W. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of the heme d1biosynthesis-associated small c-type cytochrome NirC reveals mixed oligomeric states in crystallo.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TCB
 
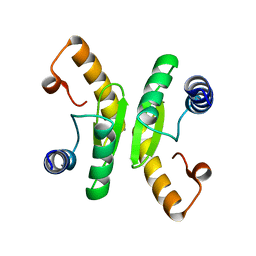 | |
6TD9
 
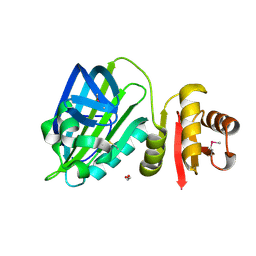 | | X-ray structure of mature PA1624 from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PA1624, ... | | Authors: | Feiler, C.G, Blankenfeldt, W. | | Deposit date: | 2019-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hypothetical periplasmic protein PA1624 from Pseudomonas aeruginosa folds into a unique two-domain structure.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TV2
 
 | | Heme d1 biosynthesis associated Protein NirF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Protein NirF, ... | | Authors: | Kluenemann, T, Layer, G, Blankenfeldt, W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Crystal structure of NirF: insights into its role in heme d 1 biosynthesis.
Febs J., 288, 2021
|
|
3REA
 
 | |
4HMV
 
 | | Crystal structure of PhzG from Pseudomonas fluorescens 2-79 in complex with tetrahydrophenazine-1-carboxylic acid after 5 days of soaking | | Descriptor: | (1R,10aS)-1,2,10,10a-tetrahydrophenazine-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, ... | | Authors: | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMW
 
 | | Crystal structure of PhzG from Burkholderia lata 383 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Pyridoxamine 5'-phosphate oxidase | | Authors: | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
