3PZ4
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BMS3 and lipid substrate FPP | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ1
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS3 | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ3
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS-analogue 14 | | Descriptor: | 4-({(3R)-7-(5-formylfuran-2-yl)-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl benzylcarbamate, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ2
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS3 and lipid substrate GGPP | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, CALCIUM ION, GERANYLGERANYL DIPHOSPHATE, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3EUV
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10, W102T, Y154T) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
3EU5
 
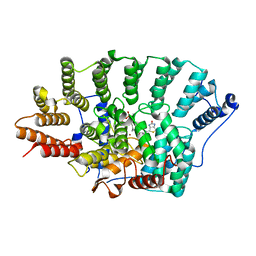 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
6ET2
 
 | |
6ET0
 
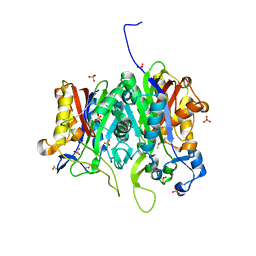 | | Crystal structure of PqsBC (C129A) mutant from Pseudomonas aeruginosa (crystal form 1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Witzgall, F, Blankenfeldt, W. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 19, 2018
|
|
6ESZ
 
 | |
3R74
 
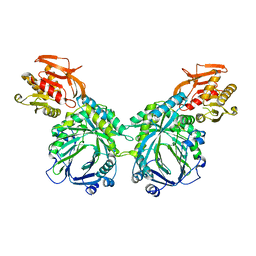 | | Crystal structure of 2-amino-2-desoxyisochorismate synthase (ADIC) synthase PhzE from Burkholderia lata 383 | | Descriptor: | Anthranilate/para-aminobenzoate synthases component I | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
3R77
 
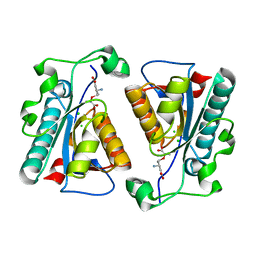 | | Crystal structure of the D38A mutant of isochorismatase PhzD from Pseudomonas fluorescens 2-79 in complex with 2-amino-2-desoxyisochorismate ADIC | | Descriptor: | (5S,6S)-6-amino-5-[(1-carboxyethenyl)oxy]cyclohexa-1,3-diene-1-carboxylic acid, CHLORIDE ION, Probable isochorismatase | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
3N6O
 
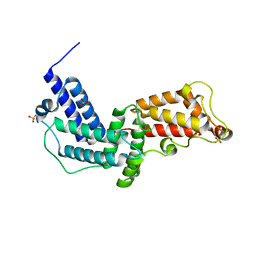 | | Crystal structure of the GEF and P4M domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | SULFATE ION, guanine nucleotide exchange factor | | Authors: | Schoebel, S, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA.
Embo Rep., 11, 2010
|
|
3NKU
 
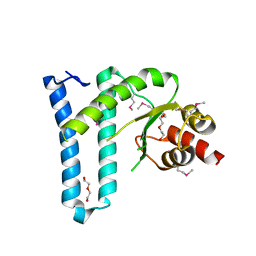 | | Crystal structure of the N-terminal domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, DrrA, TRIETHYLENE GLYCOL | | Authors: | Mueller, M.P, Peters, H, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b.
Science, 329, 2010
|
|
3NKV
 
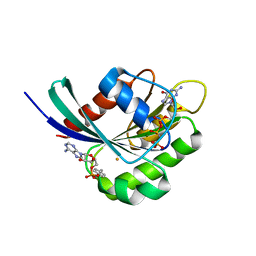 | | Crystal structure of Rab1b covalently modified with AMP at Y77 | | Descriptor: | ADENOSINE MONOPHOSPHATE, BARIUM ION, MAGNESIUM ION, ... | | Authors: | Mueller, M.P, Peters, H, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b.
Science, 329, 2010
|
|
5HWO
 
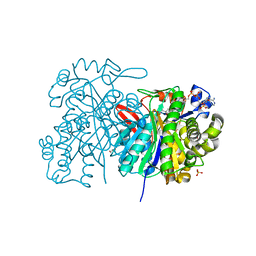 | | MvaS in complex with 3-hydroxy-3-methylglutaryl coenzyme A | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5HWP
 
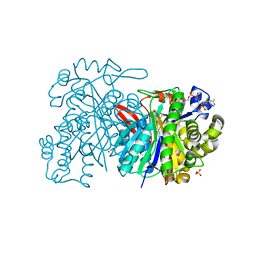 | | MvaS with acetylated Cys115 in complex with coenzyme A | | Descriptor: | COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5HWR
 
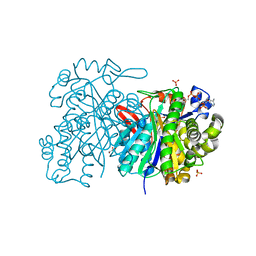 | | MvaS in complex with coenzyme A | | Descriptor: | COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5HWQ
 
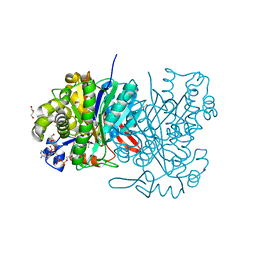 | | MvaS in complex with acetoacetyl coenzyme A | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5JBX
 
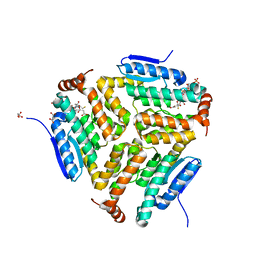 | | Crystal structure of LiuC in complex with coenzyme A and malonic acid | | Descriptor: | 3-hydroxybutyryl-CoA dehydratase, COENZYME A, MALONATE ION | | Authors: | Bock, T, Reichelt, J, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-04-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Structure of LiuC, a 3-Hydroxy-3-Methylglutaconyl CoA Dehydratase Involved in Isovaleryl-CoA Biosynthesis in Myxococcus xanthus, Reveals Insights into Specificity and Catalysis.
Chembiochem, 17, 2016
|
|
3JVJ
 
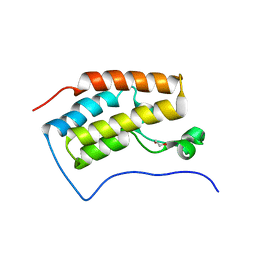 | | Crystal structure of the bromodomain 1 in mouse Brd4 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
3JVM
 
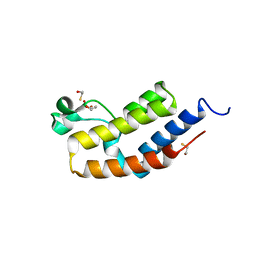 | | Crystal structure of bromodomain 2 of mouse Brd4 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromodomain-containing protein 4 | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
3DH8
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | FE (III) ION, Uncharacterized protein PA1000, bis(4-nitrophenyl) hydrogen phosphate | | Authors: | Yu, S, Blankenfeldt, W. | | Deposit date: | 2008-06-17 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
3JZA
 
 | | Crystal structure of human Rab1b in complex with the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | PHOSPHATE ION, Ras-related protein Rab-1B, Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
3JVL
 
 | | Crystal structure of bromodomain 2 of mouse Brd4 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromodomain-containing protein 4 | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
3JVK
 
 | | Crystal structure of bromodomain 1 of mouse Brd4 in complex with histone H3-K(ac)14 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, histone H3.3 peptide | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
