9F96
 
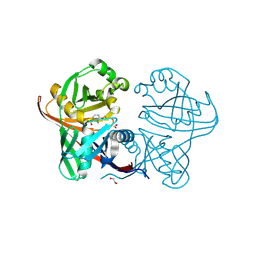 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-ethoxybenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-3-ethoxy-benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F92
 
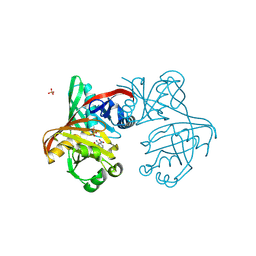 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-nitrobenzoic acid | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-azanyl-3-nitro-benzoic acid, SULFATE ION, ... | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F95
 
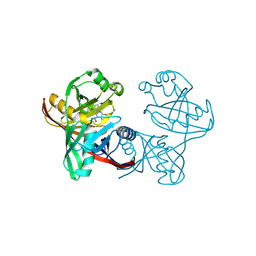 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-3-hydroxy-5-(3-hydroxyphenyl)benzoic acid | | Descriptor: | 2-azanyl-5-(3-hydroxyphenyl)-3-oxidanyl-benzoic acid, Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
9F94
 
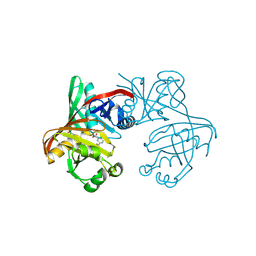 | | Complex of phenazine biosynthesis enzyme PhzF with 2-amino-5-(3-hydroxyphenyl)benzoic acid | | Descriptor: | 2-azanyl-5-(3-hydroxyphenyl)benzoic acid, ACETATE ION, FORMIC ACID, ... | | Authors: | Baumgarten, J, Schneider, P, Blankenfeldt, W, Kunick, C. | | Deposit date: | 2024-05-07 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Substrate-Based Ligand Design for Phenazine Biosynthesis Enzyme PhzF.
Chemmedchem, 2024
|
|
4X8D
 
 | | Ergothioneine-biosynthetic sulfoxide synthase EgtB in complex with N,N-dimethyl-histidine and gamma-glutamyl-cysteine | | Descriptor: | CALCIUM ION, CHLORIDE ION, GAMMA-GLUTAMYLCYSTEINE, ... | | Authors: | Vit, A, Goncharenko, K.V, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Sulfoxide Synthase EgtB from the Ergothioneine Biosynthetic Pathway.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4X8E
 
 | | Ergothioneine-biosynthetic sulfoxide synthase EgtB in complex with N,N,N-trimethyl-histidine | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Vit, A, Goncharenko, K.V, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Sulfoxide Synthase EgtB from the Ergothioneine Biosynthetic Pathway.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4AY8
 
 | | SeMet-derivative of a methyltransferase from M. mazei | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-THIOETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeldt, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3B4O
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, apo form | | Descriptor: | ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
3B4P
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, complex with 2-(cyclohexylamino)benzoic acid | | Descriptor: | 2-(cyclohexylamino)benzoic acid, ACETATE ION, AZIDE ION, ... | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
7QY3
 
 | | Crystal structure of the halohydrin dehalogenase HheG D114C mutant cross-linked with BMOE | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), Putative oxidoreductase, SULFATE ION | | Authors: | Henke, S, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Biocatalytically active and stable cross-linked enzyme crystals of halohydrin dehalogenase HheG by protein engineering
Chemcatchem, 2022
|
|
8AJQ
 
 | |
7R3J
 
 | | Nativ complex of PqsE and RhlR with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3H
 
 | |
7R3G
 
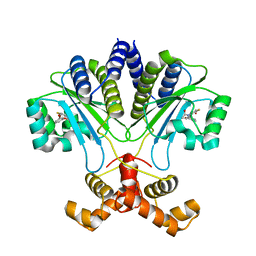 | |
7R3I
 
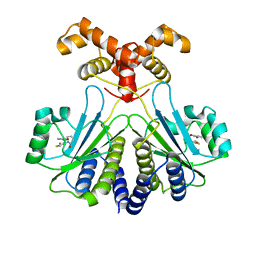 | |
4PIP
 
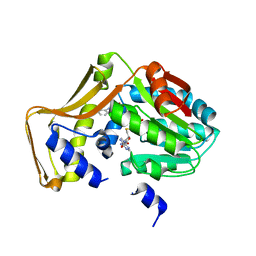 | | Engineered EgtD variant EgtD-M252V,E282A in complex with tryptophan and SAH | | Descriptor: | CHLORIDE ION, Histidine-specific methyltransferase EgtD, MAGNESIUM ION, ... | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
4X8B
 
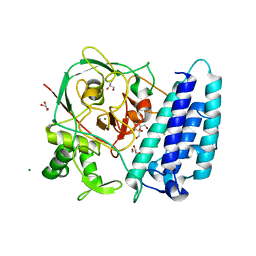 | | Ergothioneine-biosynthetic sulfoxide synthase EgtB, apo form | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Vit, A, Goncharenko, K.V, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Sulfoxide Synthase EgtB from the Ergothioneine Biosynthetic Pathway.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4PIO
 
 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with N,N-dimethylhistidine and SAH | | Descriptor: | CHLORIDE ION, Histidine-specific methyltransferase EgtD, MAGNESIUM ION, ... | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
4PIM
 
 | |
7R3F
 
 | | Monomeric PqsE mutant E187R | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, BENZOIC ACID, CACODYLATE ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3E
 
 | | Fusion construct of PqsE and RhlR in complex with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase,Regulatory protein RhlR, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
4ZFK
 
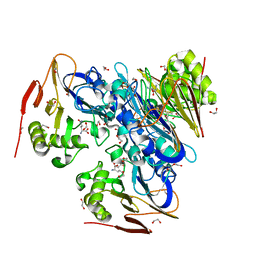 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC with glutamine | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase EgtC, GLUTAMINE | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
4ZFJ
 
 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC, apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Amidohydrolase EgtC | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
4PIN
 
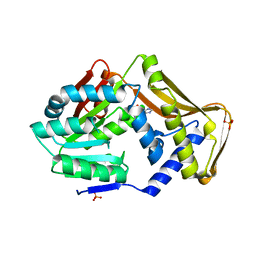 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with N,N-dimethylhistidine | | Descriptor: | Histidine-specific methyltransferase EgtD, N,N-dimethyl-L-histidine, PHOSPHATE ION | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
7ZPN
 
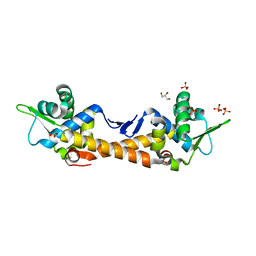 | | Crystal Structure of IscR from Dinoroseobacter shibae | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator, SULFATE ION | | Authors: | Lukat, P, Ploetzky, L, Blankenfeldt, W, Jahn, D, Haertig, E. | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dinoroseobacter shibae IscR homolog acts as a repressor for iron acquisition genes
To Be Published
|
|
