1MB4
 
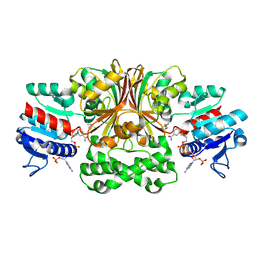 | | Crystal structure of aspartate semialdehyde dehydrogenase from vibrio cholerae with NADP and S-methyl-l-cysteine sulfoxide | | Descriptor: | Aspartate-Semialdehyde Dehydrogenase, CYSTEINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Blanco, J, Moore, R.A, Kabaleeswaran, V, Viola, R.E. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A structural Basis for the Mechanism of Aspartate-beta-semialdehyde Dehydrogenase from Vibrio Cholerae
Protein Sci., 12, 2003
|
|
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
5ET4
 
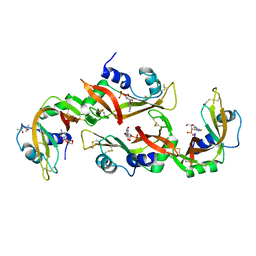 | | Structure of RNase A-K7H/R10H in complex with 3'-CMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-3'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Blanco, J.A, Salazar, V.A, Moussaoui, M, Boix, E. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
4OXB
 
 | |
4OXF
 
 | |
4OWZ
 
 | |
5OGH
 
 | | Structure of RNase A at high resolution (1.16 A) in complex with 3'-CMP and sulphate ions | | Descriptor: | CHLORIDE ION, CYTIDINE-3'-MONOPHOSPHATE, Ribonuclease pancreatic, ... | | Authors: | Blanco, J.A, Prats-Ejarque, G, Salazar, V.A, Moussaoui, M, Boix, E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
1PQU
 
 | | Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with NADP, S-methyl cysteine sulfoxide and cacodylate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CACODYLATE ION, CYSTEINE, ... | | Authors: | Blanco, J, Moore, R.A, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1MC4
 
 | |
1NWH
 
 | |
1NWC
 
 | |
1PR3
 
 | | Crystal Structure of the R103K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PU2
 
 | | Crystal Structure of the K246R Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-23 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NX6
 
 | |
1Q2X
 
 | | Crystal Structure of the E243D Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae bound with substrate aspartate semialdehyde | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PQP
 
 | | Crystal Structure of the C136S Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with Aspartate Semialdehyde and Phosphate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, L-HOMOSERINE, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Viola, R.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PS8
 
 | | Crystal Structure of the R270K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OZA
 
 | |
6B5B
 
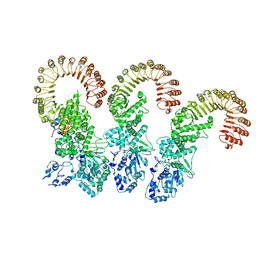 | | Cryo-EM structure of the NAIP5-NLRC4-flagellin inflammasome | | Descriptor: | Baculoviral IAP repeat-containing protein 1e, Flagellin, NLR family CARD domain-containing protein 4 | | Authors: | Tenthorey, J.L, Haloupek, N, Lopez-Blanco, J.R, Grob, P, Adamson, E, Hartenian, E, Lind, N.A, Bourgeois, N.M, Chacon, P, Nogales, E, Vance, R.E. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion.
Science, 358, 2017
|
|
8QTZ
 
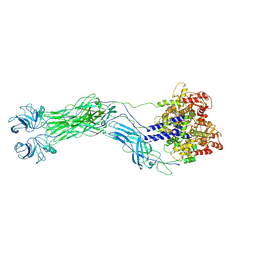 | | Cryo-EM reconstruction of VP5*/VP8* assembly from SA11 Rotavirus Tripsinized Triple Layered Particle | | Descriptor: | Outer capsid protein VP4 | | Authors: | Asensio-Cob, D, Perez-Mata, C, Gomez-Blanco, J, Vargas, J, Rodriguez, J.M, Luque, D. | | Deposit date: | 2023-10-13 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of rotavirus spike proteolytic activation
To Be Published
|
|
6QI5
 
 | | Near Atomic Structure of an Atadenovirus Shows a possible gene duplication event and Intergenera Variations in Cementing Proteins | | Descriptor: | Hexon protein, PIIIa, Penton protein, ... | | Authors: | Condezo, G.N, Marabini, R, Gomez-Blanco, J, SanMartin, C. | | Deposit date: | 2019-01-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins.
Sci Adv, 7, 2021
|
|
5FUR
 
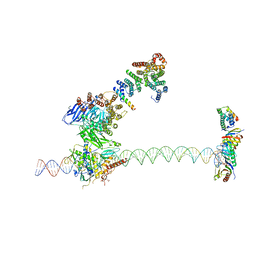 | | Structure of human TFIID-IIA bound to core promoter DNA | | Descriptor: | SUPER CORE PROMOTER, TATA-BOX-BINDING PROTEIN, TRANSCRIPTION INITIATION FACTOR IIA SUBUNIT 1, ... | | Authors: | Louder, R.K, He, Y, Lopez-Blanco, J.R, Fang, J, Chacon, P, Nogales, E. | | Deposit date: | 2016-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of Promoter-Bound TFIID and Model of Human Pre-Initiation Complex Assembly.
Nature, 531, 2016
|
|
3ZUE
 
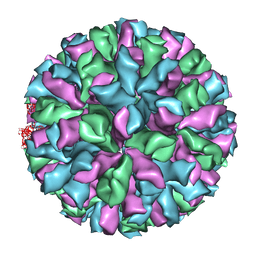 | | Rabbit Hemorrhagic Disease Virus (RHDV)capsid protein | | Descriptor: | CAPSID STRUCTURAL PROTEIN VP60 | | Authors: | Luque, D, Gonzalez, J.M, Gomez-Blanco, J, Marabini, R, Chichon, J, Mena, I, Angulo, I, Carrascosa, J.L, Verdaguer, N, Trus, B.L, Barcena, J, Caston, J.R. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Epitope Insertion at the N-Terminal Molecular Switch of the Rabbit Hemorrhagic Disease Virus T=3 Capsid Protein Leads to Larger T=4 Capsids.
J.Virol., 86, 2012
|
|
6W7W
 
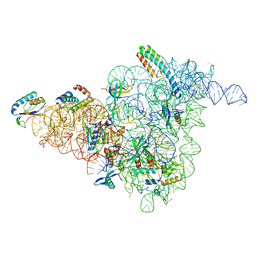 | | 30S-Inactive-low-Mg2+ Class B | | Descriptor: | 16S rRNA, 30S ribosomal protein S12, 30S ribosomal protein S15, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W6K
 
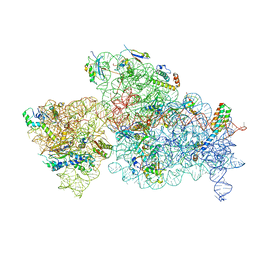 | | 30S-Activated-high-Mg2+ | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
