4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
5OGH
 
 | | Structure of RNase A at high resolution (1.16 A) in complex with 3'-CMP and sulphate ions | | Descriptor: | CHLORIDE ION, CYTIDINE-3'-MONOPHOSPHATE, Ribonuclease pancreatic, ... | | Authors: | Blanco, J.A, Prats-Ejarque, G, Salazar, V.A, Moussaoui, M, Boix, E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
4OXF
 
 | |
5ET4
 
 | | Structure of RNase A-K7H/R10H in complex with 3'-CMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-3'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Blanco, J.A, Salazar, V.A, Moussaoui, M, Boix, E. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
4OXB
 
 | |
4OWZ
 
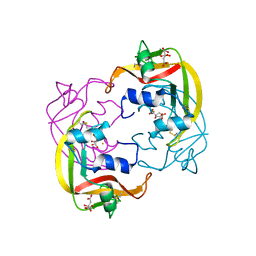 | |
1PQU
 
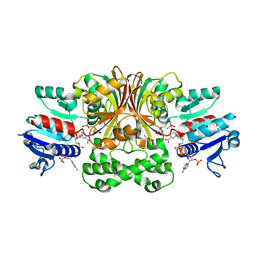 | | Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with NADP, S-methyl cysteine sulfoxide and cacodylate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CACODYLATE ION, CYSTEINE, ... | | Authors: | Blanco, J, Moore, R.A, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NX6
 
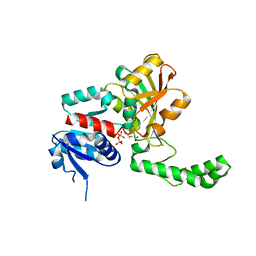 | |
1NWC
 
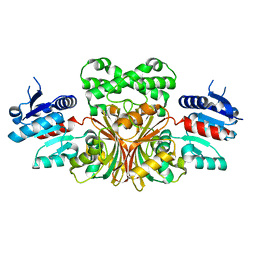 | |
1MB4
 
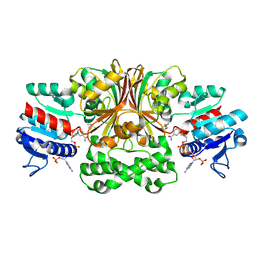 | | Crystal structure of aspartate semialdehyde dehydrogenase from vibrio cholerae with NADP and S-methyl-l-cysteine sulfoxide | | Descriptor: | Aspartate-Semialdehyde Dehydrogenase, CYSTEINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Blanco, J, Moore, R.A, Kabaleeswaran, V, Viola, R.E. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A structural Basis for the Mechanism of Aspartate-beta-semialdehyde Dehydrogenase from Vibrio Cholerae
Protein Sci., 12, 2003
|
|
1NWH
 
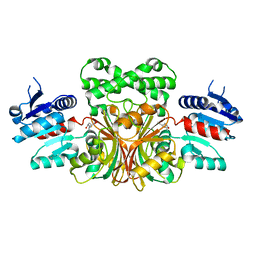 | |
1MC4
 
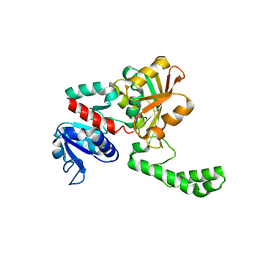 | |
1OZA
 
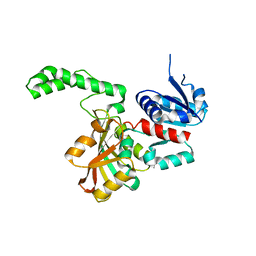 | |
1PU2
 
 | | Crystal Structure of the K246R Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-23 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Q2X
 
 | | Crystal Structure of the E243D Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae bound with substrate aspartate semialdehyde | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PQP
 
 | | Crystal Structure of the C136S Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with Aspartate Semialdehyde and Phosphate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, L-HOMOSERINE, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Viola, R.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PS8
 
 | | Crystal Structure of the R270K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PR3
 
 | | Crystal Structure of the R103K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4P7E
 
 | | Triazolopyridine compounds as selective JAK1 inhibitors: from hit identification to GLPG0634 | | Descriptor: | N-(5-{4-[(1,1-dioxidothiomorpholin-4-yl)methyl]phenyl}[1,2,4]triazolo[1,5-a]pyridin-2-yl)cyclopropanecarboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Menet, C.C.J, Fletcher, S, Van Lommen, G, Geney, R, Blanc, J, Smits, K, Jouannigot, N, van der Aar, E.M, Clement-Lacroix, P, Lepescheux, L, Galien, R, Vayssiere, B, Nelles, L, Christophe, T, Brys, R, Uhring, M, Ciesielski, F, Van Rompaey, L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triazolopyridines as Selective JAK1 Inhibitors: From Hit Identification to GLPG0634.
J.Med.Chem., 57, 2014
|
|
4ZIU
 
 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning the residues from domain MG7 to the C-terminus. | | Descriptor: | GLYCEROL, NICKEL (II) ION, Uncharacterized lipoprotein YfhM | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZJH
 
 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning domains NIE-MG1. | | Descriptor: | ACETATE ION, GLYCEROL, alpha-2-Macroglobulin | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1G2O
 
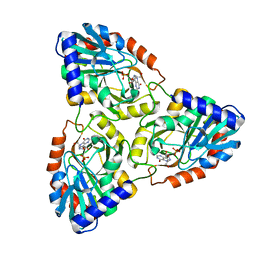 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
2Q9J
 
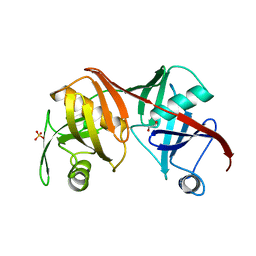 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2PR2
 
 | |
3TDT
 
 | |
