1UZR
 
 | | Crystal Structure of the Class Ib Ribonucleotide Reductase R2F-2 subunit from Mycobacterium tuberculosis | | Descriptor: | CITRIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Uppsten, M, Davis, J, Rubin, H, Uhlin, U. | | Deposit date: | 2004-03-15 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Biologically Active Form of Class Ib Ribonucleotide Reductase Small Subunit from Mycobacterium Tuberculosis
FEBS Lett., 569, 2004
|
|
4P7Q
 
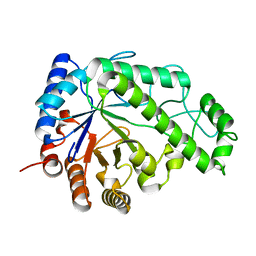 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7R
 
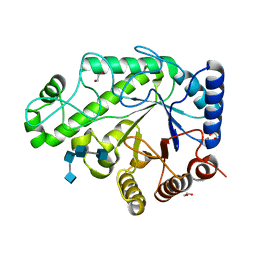 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7N
 
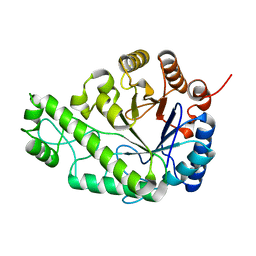 | | Structure of Escherichia coli PgaB C-terminal domain in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1FDG
 
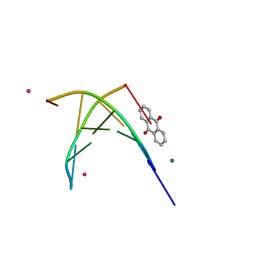 | | BINDING OF A MACROCYCLIC BISACRIDINE AND AMETANTRONE TO CGTACG INVOLVES SIMILAR UNUSUAL INTERCALATION PLATFORMS (AMETANTRONE COMPLEX) | | Descriptor: | 1,4-BIS-[2-(2-HYDROXY-ETHYLAMINO)-ETHYLAMINO]-ANTHRAQUINONE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Yang, X.-L, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of a macrocyclic bisacridine and ametantrone to CGTACG involves similar unusual intercalation platforms.
Biochemistry, 39, 2000
|
|
1VAQ
 
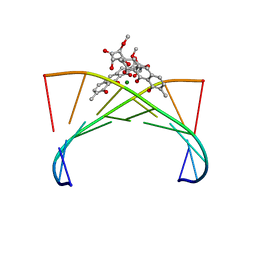 | | Crystal structure of the Mg2+-(chromomycin A3)2-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by metal ion | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Robinson, H, Gao, Y.G, Wang, A.H.-J. | | Deposit date: | 2004-02-19 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the [Mg2+-(chromomycin A3)2]-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by a metal ion
Nucleic Acids Res., 32, 2004
|
|
1U58
 
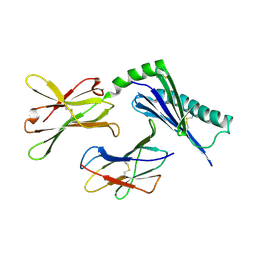 | | Crystal structure of the murine cytomegalovirus MHC-I homolog m144 | | Descriptor: | MHC-I homolog m144, beta-2-microglobulin | | Authors: | Natarajan, K, Hicks, A, Robinson, H, Guan, R, Margulies, D.H. | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the murine cytomegalovirus MHC-I homolog m144.
J.Mol.Biol., 358, 2006
|
|
1XSD
 
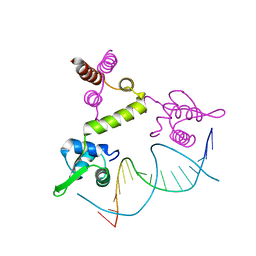 | | Crystal structure of the BlaI repressor in complex with DNA | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', penicillinase repressor | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Robinson, H, Scarsdale, N, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2004-10-19 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
4P7O
 
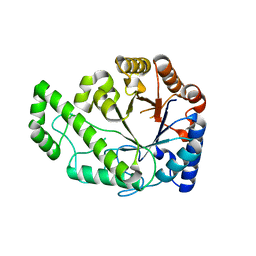 | | Structure of Escherichia coli PgaB C-terminal domain, P1 crystal form | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OZX
 
 | |
3F99
 
 | | W354F Yersinia enterocolitica PTPase apo form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase yopH | | Authors: | Brandao, T.A.S, Robinson, H, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2008-11-13 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Impaired acid catalysis by mutation of a protein loop hinge residue in a YopH mutant revealed by crystal structures.
J.Am.Chem.Soc., 131, 2009
|
|
3F9A
 
 | | W354F Yersinia enterocolitica PTPase complexed with tungstate | | Descriptor: | TUNGSTATE(VI)ION, Tyrosine-protein phosphatase yopH | | Authors: | Brandao, T.A.S, Robinson, H, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2008-11-13 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Impaired acid catalysis by mutation of a protein loop hinge residue in a YopH mutant revealed by crystal structures.
J.Am.Chem.Soc., 131, 2009
|
|
3F9B
 
 | | W354F Yersinia enterocolitica PTPase complexed with divanadate | | Descriptor: | Divanadate ion, GLYCEROL, Tyrosine-protein phosphatase yopH, ... | | Authors: | Brandao, T.A.S, Robinson, H, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2008-11-13 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Impaired acid catalysis by mutation of a protein loop hinge residue in a YopH mutant revealed by crystal structures.
J.Am.Chem.Soc., 131, 2009
|
|
3EZQ
 
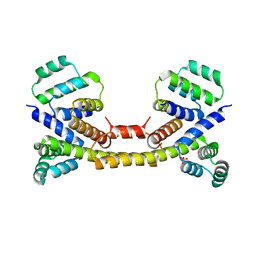 | | Crystal Structure of the Fas/FADD Death Domain Complex | | Descriptor: | Protein FADD, SODIUM ION, SULFATE ION, ... | | Authors: | Schwarzenbacher, R, Robinson, H, Stec, B, Riedl, S.J. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Nature, 457, 2009
|
|
3FVU
 
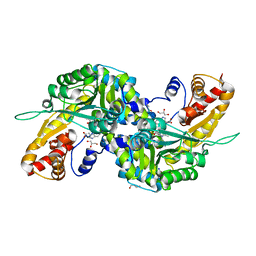 | | Crystal Structure of Human Kynurenine Aminotransferase I in Complex with Indole-3-acetic Acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, GLYCEROL, Kynurenine--oxoglutarate transaminase 1, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3FVS
 
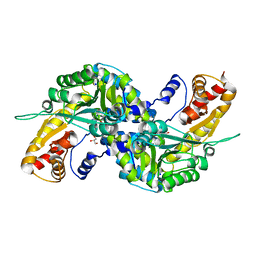 | | Human Kynurenine Aminotransferase I in complex with Glycerol | | Descriptor: | GLYCEROL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3FVX
 
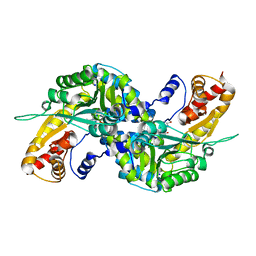 | | Human kynurenine aminotransferase I in complex with tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3IN0
 
 | |
3IN2
 
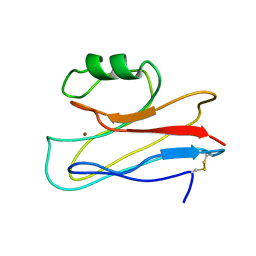 | |
3GMT
 
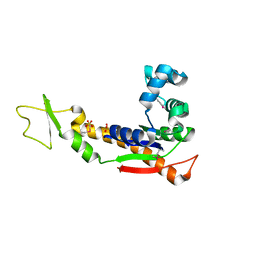 | | Crystal structure of adenylate kinase from burkholderia pseudomallei | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Abendroth, J, Staker, B.L, Robinson, H, Buchko, G.W, Hewitt, S.N, Napuli, A.J, Van Voorhis, W, Stacy, R, Myler, P.J, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-03-15 | | Release date: | 2009-06-02 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of Burkholderia pseudomallei adenylate kinase (Adk): profound asymmetry in the crystal structure of the 'open' state.
Biochem.Biophys.Res.Commun., 394, 2010
|
|
3JYB
 
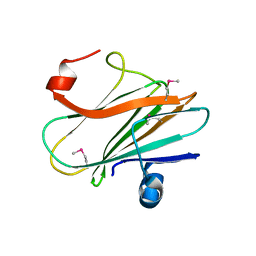 | |
3TUY
 
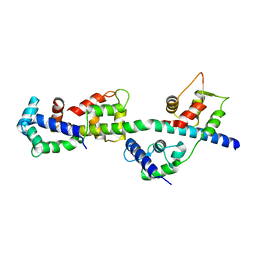 | | Phosphorylated Light Chain Domain of Scallop smooth Muscle Myosin | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Kumar, V.S.S, O'Neall-hennessey, E, Reshetnikova, L, Brown, J.H, Robinson, H, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2011-09-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a phosphorylated light chain domain of scallop smooth-muscle Myosin.
Biophys.J., 101, 2011
|
|
1M6H
 
 | | Human glutathione-dependent formaldehyde dehydrogenase | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-16 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1MA0
 
 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with NAD+ and dodecanoic acid | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, LAURIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
3T6G
 
 | |
