4LPQ
 
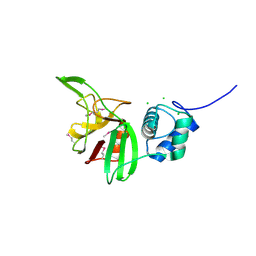 | | Crystal structure of the L,D-transpeptidase (residues 123-326) from Xylanimonas cellulosilytica DSM 15894 | | Descriptor: | CHLORIDE ION, ErfK/YbiS/YcfS/YnhG family protein | | Authors: | Nocek, B, Bigelow, L, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-16 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of the L,D-transpeptidase (residues 123-326) from Xylanimonas cellulosilytica DSM 15894
To be Published
|
|
5ULB
 
 | |
5UID
 
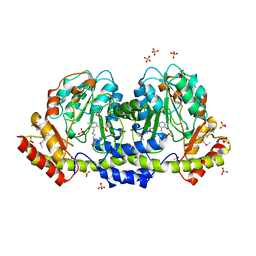 | | The crystal structure of an aminotransferase TlmJ from Streptoalloteichus hindustanus | | Descriptor: | Aminotransferase TlmJ, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Bearden, J, Phillips Jr, G.N, Joachmiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The crystal structure of an aminotransferase TlmJ from Streptoalloteichus hindustanus.
To Be Published
|
|
5UC0
 
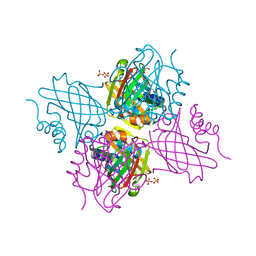 | | Crystal Structure of Beta-barrel-like, Uncharacterized Protein of COG5400 from Brucella abortus | | Descriptor: | CHLORIDE ION, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-08 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Periplasmic protein EipA determines envelope stress resistance and virulence in Brucella abortus.
Mol. Microbiol., 2018
|
|
4RGR
 
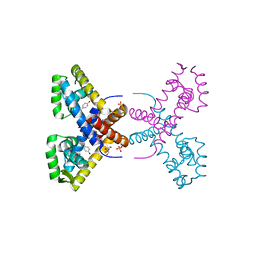 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, GLYCEROL, Repressor protein, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP
To be Published, 2014
|
|
4RGS
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with Vanilin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, GLYCEROL, Repressor protein, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with Vanilin
To be Published, 2014
|
|
4RGU
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with ferulic acid
To be Published, 2014
|
|
4RGX
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 3,4-dihydroxy bezoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 3,4-dihydroxy bezoic acid
To be Published, 2014
|
|
4ZNM
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | Descriptor: | GLYCEROL, Putative aminotransferase | | Authors: | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
4ZXW
 
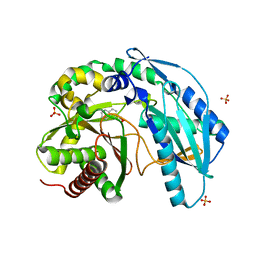 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | Descriptor: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
5BMQ
 
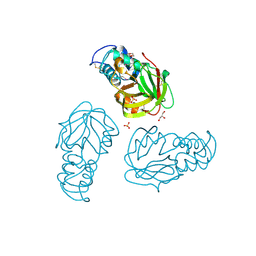 | | Crystal structure of L,D-transpeptidase (Yku) from Stackebrandtia nassauensis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ErfK/YbiS/YcfS/YnhG family protein, GLYCEROL, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of L,D-transpeptidases (Yku)
To be Published
|
|
3TEB
 
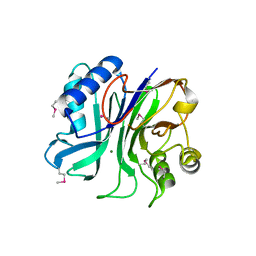 | | endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b | | Descriptor: | Endonuclease/exonuclease/phosphatase, MAGNESIUM ION | | Authors: | Chang, C, Bigelow, L, Muniez, I, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-12 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b
To be Published
|
|
3UO2
 
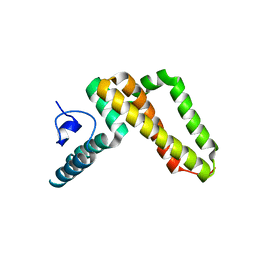 | | Jac1 co-chaperone from Saccharomyces cerevisiae | | Descriptor: | J-type co-chaperone JAC1, mitochondrial | | Authors: | Osipiuk, J, Mulligan, R, Bigelow, L, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
3T8K
 
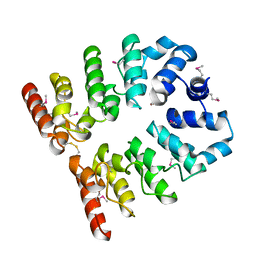 | |
3UO3
 
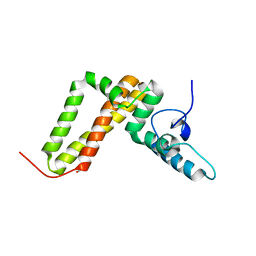 | | Jac1 co-chaperone from Saccharomyces cerevisiae, 5-182 clone | | Descriptor: | ACETATE ION, J-type co-chaperone JAC1, mitochondrial | | Authors: | Osipiuk, J, Bigelow, L, Mulligan, R, Feldmann, B, Babnigg, G, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
3U1D
 
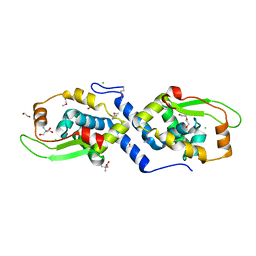 | | The structure of a protein with a GntR superfamily winged-helix-turn-helix domain from Halomicrobium mukohataei. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a protein with a GntR superfamily winged-helix-turn-helix domain from Halomicrobium mukohataei.
TO BE PUBLISHED
|
|
3U4Y
 
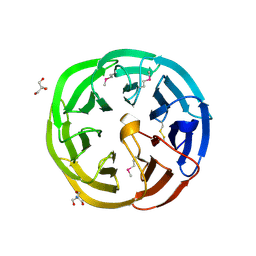 | | The crystal structure of a functionally unknown protein (Dtox_1751) from Desulfotomaculum acetoxidans DSM 771. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, uncharacterized protein | | Authors: | Tan, K, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-10 | | Release date: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | The crystal structure of a functionally unknown protein (Dtox_1751) from Desulfotomaculum acetoxidans DSM 771.
To be Published
|
|
3HM2
 
 | | Crystal Structure of Putative Precorrin-6Y C5,15-Methyltransferase Targeted Domain from Corynebacterium diphtheriae | | Descriptor: | ACETIC ACID, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Kim, Y, Bigelow, L, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-28 | | Release date: | 2009-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Crystal Structure of Putative Precorrin-6Y C5,15-Methyltransferase Targeted Domain from Corynebacterium diphtheriae
To be Published
|
|
3LFR
 
 | |
3LUY
 
 | |
2NR7
 
 | | Structural Genomics, the crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83 | | Descriptor: | Secretion activator protein, putative | | Authors: | Tan, K, Bigelow, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-01 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83
To be Published
|
|
2OKU
 
 | |
2O3F
 
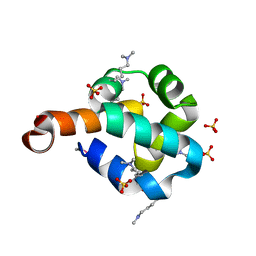 | | Structural Genomics, the crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168. | | Descriptor: | Putative HTH-type transcriptional regulator ybbH, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
2OBB
 
 | |
