6NYE
 
 | |
6NZ3
 
 | |
9EWK
 
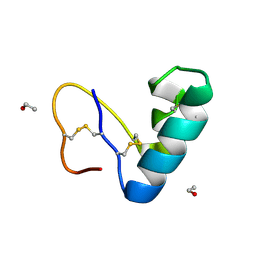 | | Solvent organization in ultrahigh-resolution protein crystal structure at room temperature | | Descriptor: | Crambin, ETHANOL | | Authors: | Chen, J.C.-H, Gilski, M, Chang, C, Borek, D, Rosenbaum, G, Lavens, A, Otwinowski, Z, Kubicki, M, Dauter, Z, Jaskolski, M, Joachimiak, A. | | Deposit date: | 2024-04-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Solvent organization in the ultrahigh-resolution crystal structure of crambin at room temperature.
Iucrj, 11, 2024
|
|
2G7L
 
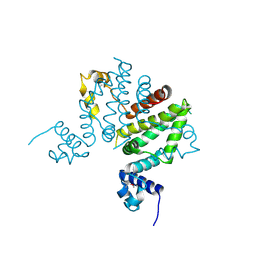 | | Crystal structure of putative transcription regulator SCO7704 from Streptomyces coelicor | | Descriptor: | TetR-family transcriptional regulator | | Authors: | Ezersky, A, Lunin, V.V, Skarina, T, Wierzbicka, M, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative transcription regulator SCO7704 from Streptomyces coelicor
To be Published
|
|
3P4J
 
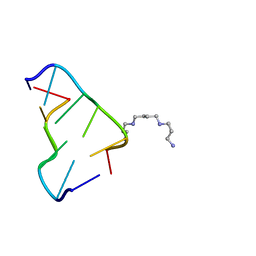 | | Ultra-high resolution structure of d(CGCGCG)2 Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE | | Authors: | Brzezinski, K, Brzuszkiewicz, A, Dauter, M, Kubicki, M, Jaskolski, M, Dauter, Z. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.55 Å) | | Cite: | High regularity of Z-DNA revealed by ultra high-resolution crystal structure at 0.55 A.
Nucleic Acids Res., 39, 2011
|
|
2N9H
 
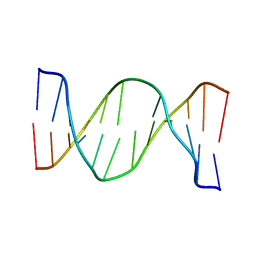 | | Glucose as a nuclease mimic in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2N9F
 
 | | Glucose as non natural nucleobase | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*(4JA)P*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6E00
 
 | | Structure of a N-Me-p-iodo-D-Phe1,N-Me-D-Gln4,Lys10-teixobactin analogue | | Descriptor: | N-Me-p-iodo-D-Phe1,N-Me-D-Gln4,Lys10-teixobactin analogue, SULFATE ION | | Authors: | Nowick, J.S, Yang, H, Wierzbicki, M. | | Deposit date: | 2018-07-05 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystallographic Structure of a Teixobactin Derivative Reveals Amyloid-like Assembly.
J. Am. Chem. Soc., 140, 2018
|
|
