5DCC
 
 | | X-RAY CRYSTAL STRUCTURE OF a TEBIPENEM ADDUCT OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-23 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
3MSV
 
 | |
4TXI
 
 | | Construct of MICAL-1 containing the monooxygenase and calponin homology domains | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alqassim, S.S, Amzel, L.M, Bianchet, M.A. | | Deposit date: | 2014-07-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Modulation of MICAL Monooxygenase Activity by its Calponin Homology Domain: Structural and Mechanistic Insights.
Sci Rep, 6, 2016
|
|
4TXK
 
 | |
1GG5
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AND A CHEMOTHERAPEUTIC DRUG (E09) AT 2.5 A RESOLUTION | | Descriptor: | 3-HYDROXYMETHYL-5-AZIRIDINYL-1METHYL-2-[1H-INDOLE-4,7-DIONE]-PROPANOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Winski, S, Hargreaves, R, Moody, C.J, Hudnott, A.R, Ross, D, Amzel, L.M. | | Deposit date: | 2000-07-12 | | Release date: | 2001-09-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based development of anticancer drugs: complexes of NAD(P)H:quinone oxidoreductase 1 with chemotherapeutic quinones.
Structure, 9, 2001
|
|
1H69
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,3,5,6,TETRAMETHYL-P-BENZOQUINONE (DUROQUINONE) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | 3-(HYDROXYMETHYL)-1-METHYL-5-(2-METHYLAZIRIDIN-1-YL)-2-PHENYL-1H-INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-08 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
1H66
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,5-diaziridinyl-3-hydroxyl-6-methyl-1,4-benzoquinone | | Descriptor: | 2,5-DIAZIRIDIN-1-YL-3-(HYDROXYMETHYL)-6-METHYLCYCLOHEXA-2,5-DIENE-1,4-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
2QR2
 
 | | HUMAN QUINONE REDUCTASE TYPE 2, COMPLEX WITH MENADIONE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MENADIONE, PROTEIN (QUINONE REDUCTASE TYPE 2), ... | | Authors: | Foster, C, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1999-04-19 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human quinone reductase type 2, a metalloflavoprotein.
Biochemistry, 38, 1999
|
|
1A78
 
 | | COMPLEX OF TOAD OVARY GALECTIN WITH THIO-DIGALACTOSE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GALECTIN-1, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Amzel, L.M, Bianchet, M.A, Ahmed, H, Vasta, G.R. | | Deposit date: | 1998-03-20 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: crystallographic studies of two protein-sugar complexes
Proteins, 40, 2000
|
|
1DXO
 
 | | Crystal structure of human NAD[P]H-QUINONE oxidoreductase CO with 2,3,5,6,tetramethyl-P-benzoquinone (duroquinone) at 2.5 Angstrom resolution | | Descriptor: | DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-12 | | Release date: | 2000-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DXQ
 
 | | CRYSTAL STRUCTURE OF MOUSE NAD[P]H-QUINONE OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1D4A
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AT 1.7 A RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1RYA
 
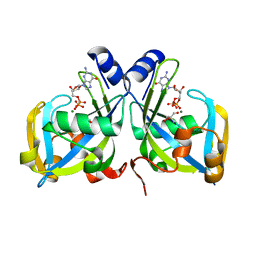 | | Crystal Structure of the E. coli GDP-mannose mannosyl hydrolase in complex with GDP and MG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GDP-mannose mannosyl hydrolase, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Legler, P.M, Mildvan, A.S, Amzel, L.M. | | Deposit date: | 2003-12-20 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of GDP-mannose glycosyl hydrolase, a Nudix enzyme that cleaves at carbon instead of phosphorus.
Structure, 12, 2004
|
|
4OVN
 
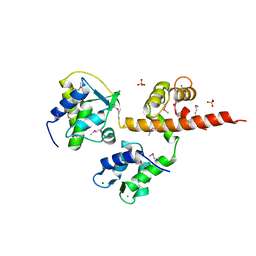 | | Voltage-gated Sodium Channel 1.5 (Nav1.5) C-terminal domain in complex with Calmodulin poised for activation | | Descriptor: | Calmodulin, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Boto, A, Jakoncic, J, Tomaselli, G.F, Amzel, L.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of the NaV1.5 cytoplasmic domain by calmodulin.
Nat Commun, 5, 2014
|
|
5D7H
 
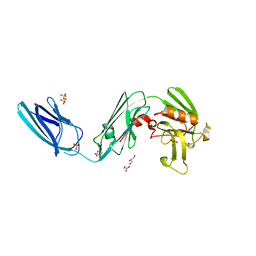 | | X-RAY CRYSTAL STRUCTURE OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | GLYCEROL, L,D-transpeptidase 2, SULFATE ION | | Authors: | Saavedra, H, Basta, L.A, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
3TX4
 
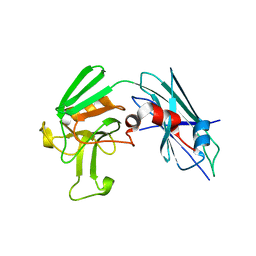 | | Crystal Structure of Mutant (C354A) M. tuberculosis LD-transpeptidase type 2 | | Descriptor: | Mycobacterium Tuberculosis LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Targeting the Cell Wall of Mycobacterium tuberculosis: Structure and Mechanism of L,D-Transpeptidase 2.
Structure, 20, 2012
|
|
3VAE
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 with Modified Catalytic Cysteine (C354) | | Descriptor: | DI(HYDROXYETHYL)ETHER, LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-12-29 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Cell Wall of Mycobacterium tuberculosis: Structure and Mechanism of L,D-Transpeptidase 2.
Structure, 20, 2012
|
|
3U1P
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 with Modified Catalytic Cysteine (C354) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Mycobacteria Tuberculosis LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-30 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Cell Wall of Mycobacterium tuberculosis: Structure and Mechanism of L,D-Transpeptidase 2.
Structure, 20, 2012
|
|
3U1Q
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 with 2-Mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, Mycobacteria Tuberculosis LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-30 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Mycobacterium tuberculosis L,D-transpeptidase 2 provides insights into targeting the cell wall of persisters
to be published
|
|
1KHZ
 
 | | Structure of the ADPR-ase in complex with AMPCPR and Mg | | Descriptor: | ADP-ribose pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2001-12-01 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase.
Biochemistry, 41, 2002
|
|
2GT4
 
 | | Crystal Structure of the Y103F mutant of the GDP-mannose mannosyl hydrolase in complex with GDP-mannose and MG+2 | | Descriptor: | GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Azurmendi, H.F, Mildvan, A.S, Amzel, L.A. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray, NMR, and mutational studies of the catalytic cycle of the GDP-mannose mannosyl hydrolase reaction.
Biochemistry, 45, 2006
|
|
2GT2
 
 | | Structure of the E. coli GDP-mannose mannosyl hydrolase | | Descriptor: | GDP-mannose mannosyl hydrolase | | Authors: | Gabelli, S.B, Bianchet, M.A, Azurmendi, H.F, MIldvan, A.S, Amzel, L.M. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, NMR, and mutational studies of the catalytic cycle of the GDP-mannose mannosyl hydrolase reaction.
Biochemistry, 45, 2006
|
|
2O1C
 
 | |
2O5W
 
 | | Structure of the E. coli dihydroneopterin triphosphate pyrophosphohydrolase in complex with Sm+3 and pyrophosphate | | Descriptor: | PYROPHOSPHATE, SAMARIUM (III) ION, SODIUM ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2006-12-06 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of the E. coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis.
Structure, 15, 2007
|
|
1GA7
 
 | | CRYSTAL STRUCTURE OF THE ADP-RIBOSE PYROPHOSPHATASE IN COMPLEX WITH GD+3 | | Descriptor: | GADOLINIUM ION, HYPOTHETICAL 23.7 KDA PROTEIN IN ICC-TOLC INTERGENIC REGION | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2000-11-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family.
Nat.Struct.Biol., 8, 2001
|
|
