4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7YK5
 
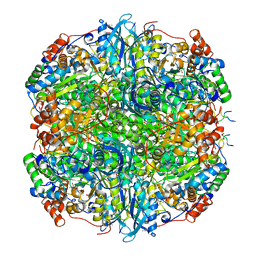 | | Rubisco from Phaeodactylum tricornutum bound to PYCO1(452-592) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, Multifunctional fusion protein, PYCO1 LSU binding motif, ... | | Authors: | Oh, Z.G, Ang, W.S.L, Bhushan, S, Mueller-Cajar, O. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | A linker protein from a red-type pyrenoid phase separates with Rubisco via oligomerizing sticker motifs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WF4
 
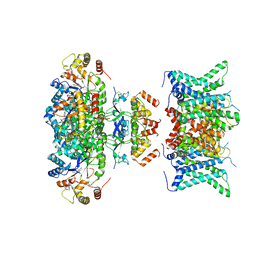 | | Composite map of human Kv1.3 channel in dalazatide-bound state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WF3
 
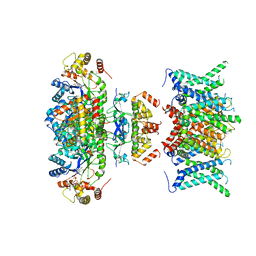 | | Composite map of human Kv1.3 channel in apo state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4V7E
 
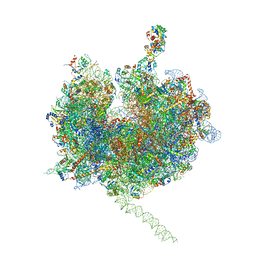 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | Authors: | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
5TCU
 
 | | Methicillin sensitive Staphylococcus aureus 70S ribosome | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Eyal, Z, Ahmed, T, Belousoff, N, Mishra, S, Matzov, D, Bashan, A, Zimmerman, E, Lithgow, T, Bhushan, S, Yonath, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome.
MBio, 8, 2017
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | Descriptor: | rRNA expansion segment ES27L in an "out" conformation | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WWA
 
 | | Cryo-EM structure of idle yeast Ssh1 complex bound to the yeast 80S ribosome | | Descriptor: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
2WWB
 
 | | CRYO-EM STRUCTURE OF THE MAMMALIAN SEC61 COMPLEX BOUND TO THE ACTIVELY TRANSLATING WHEAT GERM 80S RIBOSOME | | Descriptor: | 25S RRNA, 5.8S RRNA, 60S RIBOSOMAL PROTEIN L17-A, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.48 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
2WW9
 
 | | Cryo-EM structure of the active yeast Ssh1 complex bound to the yeast 80S ribosome | | Descriptor: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
7CBP
 
 | | CryoEM structure of Zika virus with Fab at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Fab Heavy chain, ... | | Authors: | Tyagi, A, Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2020-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A complex between the Zika virion and the Fab of a broadly cross-reactive neutralizing monoclonal antibody revealed by cryo-EM and single particle analysis at 4.1 angstrom resolution.
J Struct Biol X, 4, 2020
|
|
5FSW
 
 | | RNA dependent RNA polymerase QDE-1 from Thielavia terrestris | | Descriptor: | RNA DEPENDENT RNA POLYMERASE QDE-1 | | Authors: | Qian, X, Hamid, F.M, El Sahili, A, Darwis, D.A, Wong, Y.H, Bhushan, S, Makeyev, E.V, Lescar, J. | | Deposit date: | 2016-01-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Functional Evolution in Orthologous Cell-Encoded RNA-Dependent RNA Polymerases
J.Biol.Chem., 291, 2016
|
|
5H1S
 
 | | Structure of the large subunit of the chloro-ribosome | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L17, ... | | Authors: | Ahmed, T, Yin, Z, Bhushan, S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-01 | | Last modified: | 2018-06-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the large subunit of the spinach chloroplast ribosome.
Sci Rep, 6, 2016
|
|
5IMR
 
 | | Structure of ribosome bound to cofactor at 5.7 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5IMQ
 
 | | Structure of ribosome bound to cofactor at 3.8 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5A9Z
 
 | | Complex of Thermous thermophilus ribosome bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AA0
 
 | | Complex of Thermous thermophilus ribosome (A-and P-site tRNA) bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5ZEU
 
 | | M. smegmatis P/P state 30S ribosomal subunit | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5ZET
 
 | | M. smegmatis P/P state 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5ZEP
 
 | | M. smegmatis hibernating state 70S ribosome structure | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5ZEY
 
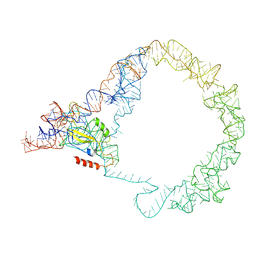 | | M. smegmatis Trans-translation state 70S ribosome | | Descriptor: | A-tRNAfMet, SsrA-binding protein, tmRNA | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5ZEB
 
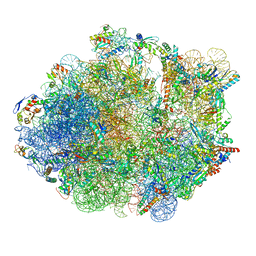 | | M. Smegmatis P/P state 70S ribosome structure | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5X8T
 
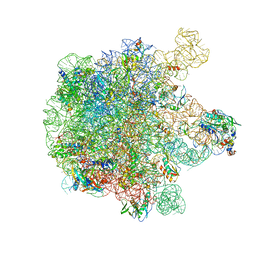 | | Structure of the 50S large subunit of chloroplast ribosome from spinach | | Descriptor: | 23S rRNA, 4.8S rRNA, 50S ribosomal protein L13, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
5X8R
 
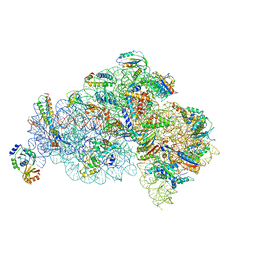 | | Structure of the 30S small subunit of chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, chloroplastic, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
5X8P
 
 | | Structure of the 70S chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
