8IL6
 
 | |
8IL1
 
 | |
5XOA
 
 | |
4PTK
 
 | | Crystal structure of Staphylococcal IMPase-I complex with 3Mg2+ and Phosphate | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2014-03-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
4RUV
 
 | | Crystal structure of thioredoxin 2 from Staphylococcus aureus NCTC8325 | | Descriptor: | Thioredoxin | | Authors: | Bose, M, Biswas, R, Roychowdhury, A, Bhattacharyya, S, Ghosh, A.K, Das, A.K. | | Deposit date: | 2014-11-22 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Elucidation of the mechanism of disulfide exchange between staphylococcal thioredoxin2 and thioredoxin reductase2: A structural insight.
Biochimie, 160, 2019
|
|
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8X3N
 
 | |
8X40
 
 | |
8XVW
 
 | | Crystal structure of N-terminal deletion mutant of Staphylococcal Thiol Peroxidase | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Shukla, M, Maji, S, Yadav, V.K, Das, A.K, Bhattacharyya, S. | | Deposit date: | 2024-01-15 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of N-terminal deletion mutant of Staphylococcal Thiol Peroxidase
To Be Published
|
|
5DW8
 
 | |
4I40
 
 | | crystal structure of Staphylococcal inositol monophosphatase-1: 50mM LiCl inhibited complex | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2012-11-27 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
4I3Y
 
 | | Crystal structure of Staphylococcal inositol monophosphatase-1: 100 mM LiCl soaked inhibitory complex | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
5I3S
 
 | |
5J16
 
 | |
3T0W
 
 | | Fluorogen activating protein M8VL in complex with dimethylindole red | | Descriptor: | 1-(3-sulfopropyl)-4-[(1E,3E)-3-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)prop-1-en-1-yl]quinolinium, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
3SJ7
 
 | |
3T0X
 
 | | Fluorogen Activating Protein M8VLA4(S55P) in complex with dimethylindole red | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-sulfopropyl)-4-[(1E,3E)-3-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)prop-1-en-1-yl]quinolinium, Immunoglobulin variable lambda domain M8VLA4(S55P), ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
3T0V
 
 | | Unliganded fluorogen activating protein M8VL | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
3T0J
 
 | |
3Q6I
 
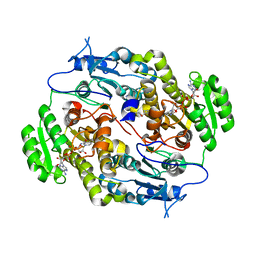 | | Crystal structure of FabG4 and coenzyme binary complex | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-01-01 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of holoFabG4
To be Published
|
|
8IKQ
 
 | |
