1JQ8
 
 | | Design of specific inhibitors of phospholipase A2: Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Leu-Ala-Ile-Tyr-Ser at 2.0 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2, ... | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of specific peptide inhibitors of phospholipase A2: structure of a complex formed between Russell's viper phospholipase A2 and a designed peptide Leu-Ala-Ile-Tyr-Ser (LAIYS).
ACTA CRYSTALLOGR.,SECT.D, 58, 2002
|
|
6Z27
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LCP crystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Betzel, C, Selikhanov, G.K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
1KPM
 
 | | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by Vitamin E and its Implications in Inflammation: Crystal Structure of the Complex Formed between Phospholipase A2 and Vitamin E at 1.8 A Resolution. | | Descriptor: | ACETIC ACID, Phospholipase A2, VITAMIN E | | Authors: | Chandra, V, Jasti, J, Kaur, P, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-01-01 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by alpha-Tocopherol (Vitamin E) and its
Implications in Inflammation: Crystal Structure of the Complex Formed Between Phospholipase A2 and
alpha-Tocopherol at 1.8 A Resolution
J.Mol.Biol., 320, 2002
|
|
6Z02
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV in surfo crystallization | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Fufina, T.Y, Vasilieva, L.G, Betzel, C. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
6Z1J
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LSP co-crystallization with spheroidene | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Betzel, C, Selikhanov, G.K. | | Deposit date: | 2020-05-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
3GVN
 
 | | The 1.2 Angstroem crystal structure of an E.coli tRNASer acceptor stem microhelix reveals two magnesium binding sites | | Descriptor: | 5'-R(*CP*CP*UP*CP*AP*CP*C)-3', 5'-R(*GP*GP*UP*GP*AP*GP*G)-3', MAGNESIUM ION | | Authors: | Eichert, A, Furste, J.P, Schreiber, A, Perbandt, M, Betzel, C, Erdmann, V.A, Forster, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2A crystal structure of an E. coli tRNASer)acceptor stem microhelix reveals two magnesium binding sites.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
5LH1
 
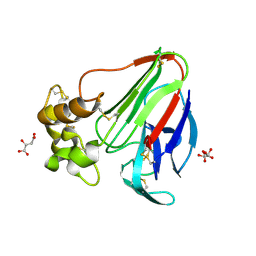 | | Low dose Thaumatin - 360-400 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
2YVV
 
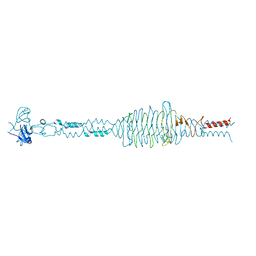 | | Crystal structure of hyluranidase complexed with lactose at 2.6 A resolution reveals three specific sugar recognition sites | | Descriptor: | Hyaluronidase, phage associated, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mishra, P, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Bhakuni, V, Singh, T.P. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of hyluranidase complexed with lactose at 2.6 A resolution reveals three specific sugar recognition sites
To be Published
|
|
3T62
 
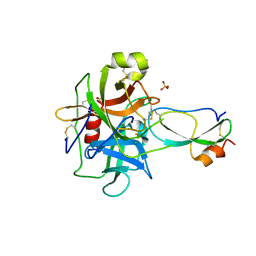 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean Sea anemone Stichodactyla helianthus in complex with bovine chymotrypsin | | Descriptor: | Chymotrypsinogen A, Kunitz-type proteinase inhibitor SHPI-1, SULFATE ION | | Authors: | Garcia-Fernandez, R, Dominguez, R, Oberthuer, D, Pons, T, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into chymotrypsin inhibition by the Kunitz-type inhibitor-1 from the marine invertebrate Stichodactyla helianthus
To be Published
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5LMH
 
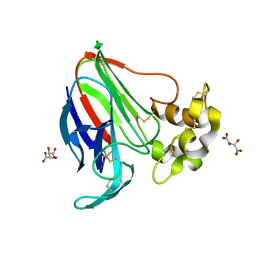 | | High dose Thaumatin - 160-200 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
1BJR
 
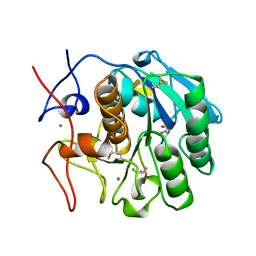 | | COMPLEX FORMED BETWEEN PROTEOLYTICALLY GENERATED LACTOFERRIN FRAGMENT AND PROTEINASE K | | Descriptor: | CALCIUM ION, LACTOFERRIN, PROTEINASE K | | Authors: | Singh, T.P, Sharma, S, Karthikeyan, S, Betzel, C, Bhatia, K.L. | | Deposit date: | 1998-06-27 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of a complex formed between proteolytically-generated lactoferrin fragment and proteinase K.
Proteins, 33, 1998
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5LH5
 
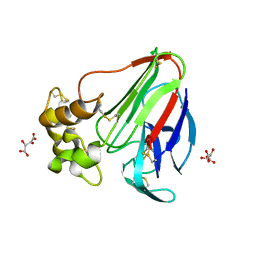 | | High dose Thaumatin - 40-80 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
6EYP
 
 | | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Prokofev, I.I, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A
To Be Published
|
|
6FJS
 
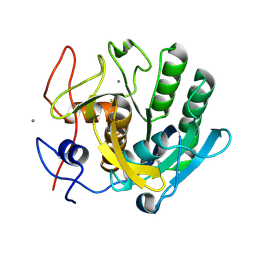 | | Proteinase~K SIRAS phased structure of room-temperature, serially collected synchrotron data | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Botha, S, Baitan, D, Jungnickel, K.E.J, Oberthuer, D, Schmidt, C, Stern, S, Wiedorn, M.O, Perbandt, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novoprotein structure determination by heavy-atom soaking in lipidic cubic phase and SIRAS phasing using serial synchrotron crystallography.
IUCrJ, 5, 2018
|
|
6Y0H
 
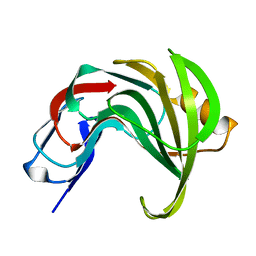 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
3UX7
 
 | | Crystal structure of a dimeric myotoxic component of the Vipera ammodytes meridionalis venom reveals determinants of myotoxicity and membrane damaging activity | | Descriptor: | Ammodytin L(1) isoform, SULFATE ION | | Authors: | Georgieva, D, Coronado, M, Oberthuer, D, Buck, F, Duhalov, D, Arni, R.K, Genov, N, Betzel, C. | | Deposit date: | 2011-12-04 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of a dimeric Ser49 PLA(2)-like myotoxic component of the Vipera ammodytes meridionalis venomics reveals determinants of myotoxicity and membrane damaging activity.
Mol Biosyst, 8, 2012
|
|
1JQ9
 
 | | Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Phe-Leu-Ser-Tyr-Lys at 1.8 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2 | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Complex Formed between a Snake Venom Phospholipase A2 and a Potent Peptide Inhibitor Phe-Leu-Ser-Tyr-Lys at 1.8 A Resolution
J.BIOL.CHEM., 277, 2002
|
|
5LH0
 
 | | Low dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH7
 
 | | High dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LN0
 
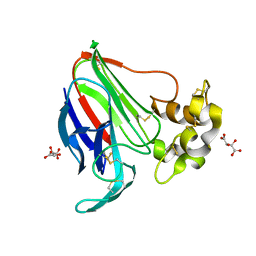 | | Low dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH3
 
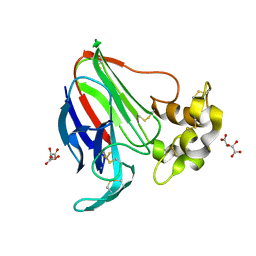 | | High dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH6
 
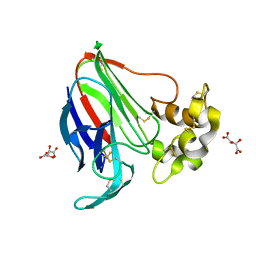 | | High dose Thaumatin - 360-400 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
3R3L
 
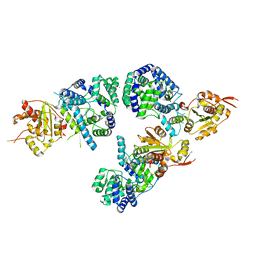 | | Structure of NP protein from Lassa AV strain | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, ZINC ION | | Authors: | Perbandt, M, Brunotte, L, Gunther, S, Betzel, C. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Structure of the Lassa virus nucleoprotein revealed by X-ray crystallography, small-angle X-ray scattering, and electron microscopy.
J.Biol.Chem., 286, 2011
|
|
