5TZN
 
 | | Structure of the viral immunoevasin m12 (Smith) bound to the natural killer cell receptor NKR-P1B (B6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein family protein m12, ... | | Authors: | Berry, R, Rossjohn, J. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Viral Immunoevasin Controls Innate Immunity by Targeting the Prototypical Natural Killer Cell Receptor Family.
Cell, 169, 2017
|
|
5J6H
 
 | |
5J6G
 
 | |
4PN6
 
 | | Structure of the Cytomegalovirus-Encoded m04 Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, M04, ... | | Authors: | Berry, R, Rossjohn, J. | | Deposit date: | 2014-05-23 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of the Cytomegalovirus-Encoded m04 Glycoprotein, a Prototypical Member of the m02 Family of Immunoevasins.
J.Biol.Chem., 289, 2014
|
|
4JO8
 
 | | Crystal structure of the activating Ly49H receptor in complex with m157 (G1F strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor 8, M157 | | Authors: | Berry, R, Ng, N, Saunders, P.M, Vivian, J.P, Lin, J, Deuss, F.A, Corbett, A.J, Forbes, C.A, Widjaja, J.M, Sullivan, L.C, McAlister, A.D, Perugini, M.A, Call, M.J, Scalzo, A.A, Degli-Esposti, M.A, Coudert, J.D, Beddoe, T, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Targeting of a natural killer cell receptor family by a viral immunoevasin
Nat.Immunol., 14, 2013
|
|
6E7D
 
 | | Structure of the inhibitory NKR-P1B receptor bound to the host-encoded ligand, Clr-b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, Killer cell lectin-like receptor subfamily B member 1B allele B, ... | | Authors: | Balaji, G.R, Rossjohn, J, Berry, R. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of host Clr-b by the inhibitory NKR-P1B receptor provides a basis for missing-self recognition.
Nat Commun, 9, 2018
|
|
6ARQ
 
 | | Crystal structure of CD96 (D1) bound to CD155/necl-5 (D1-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T-cell surface protein tactile, ... | | Authors: | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | Deposit date: | 2017-08-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155.
Structure, 27, 2019
|
|
5V52
 
 | | Structure of TIGIT bound to nectin-2 (CD112) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Nectin-2, ... | | Authors: | Deuss, F.A, Gully, B.S, Rossjohn, J, Berry, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Recognition of nectin-2 by the natural killer cell receptor T cell immunoglobulin and ITIM domain (TIGIT).
J. Biol. Chem., 292, 2017
|
|
4QRP
 
 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL and DD31 TCR | | Descriptor: | Beta-2-microglobulin, DD31 TCR alpha chain, DD31 TCR beta chain, ... | | Authors: | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
4QRQ
 
 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
7JPT
 
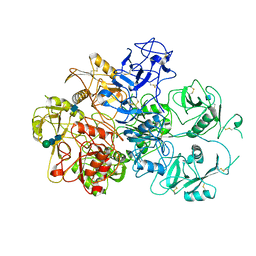 | | Structure of an endocytic receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 75, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gully, B.S, Rossjohn, J, Berry, R. | | Deposit date: | 2020-08-09 | | Release date: | 2020-12-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of the endocytic receptor DEC-205.
J.Biol.Chem., 296, 2020
|
|
7JPU
 
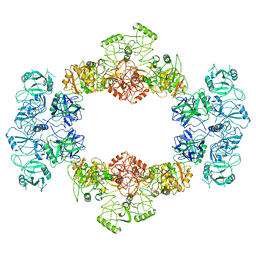 | |
2MIM
 
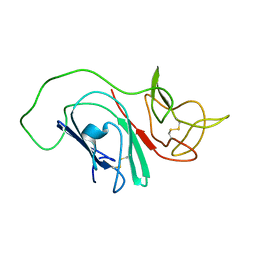 | |
8FU4
 
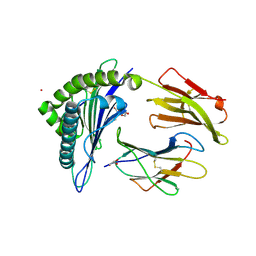 | | HCMV US11 peptide binding to HLA-A*02:01 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, GLYCEROL, ... | | Authors: | Watson, G.M, Berry, R, Littler, D.R, Rossjohn, J. | | Deposit date: | 2023-01-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diverse cytomegalovirus US11 antagonism and MHC-A evasion strategies reveal a tit-for-tat coevolutionary arms race in hominids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8FRT
 
 | | X-ray crystal structure of the N-terminal region from HCMV US11 binding to HLA-A*02:01 | | Descriptor: | GLYCEROL, Unique short US11 glycoprotein, Beta-2-microglobulin, ... | | Authors: | Watson, G.M, Berry, R, Rossjohn, J. | | Deposit date: | 2023-01-08 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse cytomegalovirus US11 antagonism and MHC-A evasion strategies reveal a tit-for-tat coevolutionary arms race in hominids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6O3O
 
 | | Structure of human DNAM-1 (CD226) bound to nectin-like protein-5 (necl-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, ... | | Authors: | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of nectin-like protein-5 by the human-activating immune receptor, DNAM-1.
J.Biol.Chem., 294, 2019
|
|
8FMA
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C11 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM9
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C12 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FMB
 
 | | Nodavirus RNA replication protein A polymerase domain, local refinement | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
