1JS7
 
 | | Solution Structure of dAAUAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*UP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1TJC
 
 | | Crystal structure of peptide-substrate-binding domain of human type I collagen prolyl 4-hydroxylase | | Descriptor: | Prolyl 4-hydroxylase alpha-1 subunit | | Authors: | Pekkala, M, Hieta, R, Bergmann, U, Kivirikko, K.I, Wierenga, R.K, Myllyharju, J. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Peptide-Substrate-binding Domain of Collagen Prolyl 4-Hydroxylases Is a Tetratricopeptide Repeat Domain with Functional Aromatic Residues.
J.Biol.Chem., 279, 2004
|
|
1D1J
 
 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
1F7S
 
 | | CRYSTAL STRUCTURE OF ADF1 FROM ARABIDOPSIS THALIANA | | Descriptor: | ACTIN DEPOLYMERIZING FACTOR (ADF), LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Bowman, G.D, Nodelman, I.M, Lindberg, U, Chua, N.H, Schutt, C.E. | | Deposit date: | 2000-06-27 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comparative structural analysis of the ADF/cofilin family.
Proteins, 41, 2000
|
|
2X80
 
 | | P450 BM3 F87A in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450/NADPH-P450 REDUCTASE, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Wong, T.S, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2010-03-05 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Role of Active-Site Phe87 in Modulating the Organic Co-Solvent Tolerance of Cytochrome P450 Bm3 Monooxygenase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2X7Y
 
 | | P450 BM3 F87A in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450/NADPH-P450 REDUCTASE, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Wong, T.S, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2010-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Role of Active-Site Phe87 in Modulating the Organic Co-Solvent Tolerance of Cytochrome P450 Bm3 Monooxygenase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3WJD
 
 | | Crystal structure of mutant nitrobindin F44W/M75L/H76L/Q96C/M148L/H158L (NB5) from Arabidopsis thaliana | | Descriptor: | GLYCEROL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WJB
 
 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/M148L/H158L (NB4) from Arabidopsis thaliana | | Descriptor: | BARIUM ION, HEXAETHYLENE GLYCOL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WJE
 
 | | Crystal structure of mutant nitrobindin M75W/H76L/Q96C/M148L/H158L (NB6) from Arabidopsis thaliana | | Descriptor: | UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WJF
 
 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/V128W/M148L/H158L (NB9) from Arabidopsis thaliana | | Descriptor: | UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WJC
 
 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/M148L/H158L covalently linked with [Rh(Cp-Mal)(COD)] (NB4-Rh) from Arabidopsis thaliana | | Descriptor: | BARIUM ION, UPF0678 fatty acid-binding protein-like protein At1g79260, [(1,2,5,6-eta)-cyclooctane-1,2,5,6-tetrayl]{(1,2,3,4,5-eta)-1-[2-(2,5-dioxopyrrolidin-1-yl)ethyl]cyclopentadienyl}rhodium | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WJG
 
 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/M148W/H158L (NB10) from Arabidopsis thaliana | | Descriptor: | GLYCEROL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
2J1M
 
 | | P450 BM3 Heme domain in complex with DMSO | | Descriptor: | CYTOCHROME P450 102, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Tuck-Seng, W, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2006-08-14 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding a Mechanism of Organic Cosolvent Inactivation in Heme Monooxygenase P450 Bm-3.
J.Am.Chem.Soc., 129, 2007
|
|
2J4S
 
 | | P450 BM3 heme domain in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Tuck-Seng, W, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2006-09-05 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding a Mechanism of Organic Cosolvent Inactivation in Heme Monooxygenase P450 Bm-3.
J.Am.Chem.Soc., 129, 2007
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
6EHH
 
 | | Crystal structure of mouse MTH1 mutant L116M with inhibitor TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gustafsson, R, Narwal, M, Jemth, A.-S, Almlof, I, Warpman Berglund, U, Helleday, T, Stenmark, P. | | Deposit date: | 2017-09-13 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
4IXR
 
 | | RT fs X-ray diffraction of Photosystem II, first illuminated state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
7ALE
 
 | | Crystal structure of human PAICS in complex with inhibitor 69 | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 2-azanyl-~{N}-[2-bromanyl-5-[4-[3-(dimethylamino)propylsulfonyl]piperazin-1-yl]phenyl]-1,3-oxazole-4-carboxamide, Multifunctional protein ADE2 | | Authors: | Skerlova, J, Marttila, P, Unterlass, J, Jemth, A.-S, Henriksson, M, Wakchaure, P, Grube, M, Warpman Berglund, U, Homan, E, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cellular and biochemical validation of a potent PAICS inhibitor
To Be Published
|
|
4IXQ
 
 | | RT fs X-ray diffraction of Photosystem II, dark state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.7 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
3NUL
 
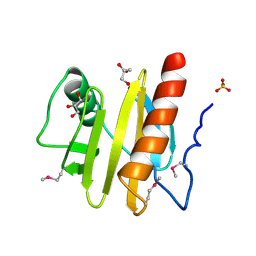 | | Profilin I from Arabidopsis thaliana | | Descriptor: | GLYCEROL, PROFILIN I, SULFATE ION | | Authors: | Thorn, K, Christensen, H.E.M, Shigeta, R, Huddler, D, Chua, N.-H, Shalaby, L, Lindberg, U, Schutt, C.E. | | Deposit date: | 1996-11-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a major allergen from plants.
Structure, 5, 1997
|
|
4HGH
 
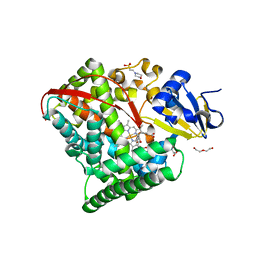 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset I) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGF
 
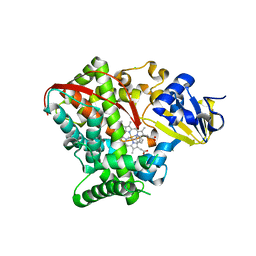 | | Crystal structure of P450 BM3 5F5K heme domain variant complexed with styrene | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGG
 
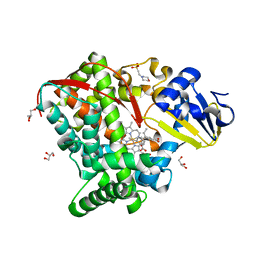 | | Crystal structure of P450 BM3 5F5R heme domain variant complexed with styrene | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGJ
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
