1NYA
 
 | | NMR SOLUTION STRUCTURE OF CALERYTHRIN, AN EF-HAND CALCIUM-BINDING PROTEIN | | Descriptor: | CALCIUM ION, Calerythrin | | Authors: | Tossavainen, H, Permi, P, Annila, A, Kilpelainen, I, Drakenberg, T. | | Deposit date: | 2003-02-12 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of calerythrin, an EF-hand calcium-binding protein from Saccharopolyspora erythraea
Eur.J.Biochem., 270, 2003
|
|
3GNU
 
 | | Toxin fold as basis for microbial attack and plant defense | | Descriptor: | 25 kDa protein elicitor, CHLORIDE ION, GUANIDINE | | Authors: | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | Deposit date: | 2009-03-18 | | Release date: | 2009-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GNZ
 
 | | Toxin fold for microbial attack and plant defense | | Descriptor: | 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | Deposit date: | 2009-03-18 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1BF9
 
 | | N-TERMINAL EGF-LIKE DOMAIN FROM HUMAN FACTOR VII, NMR, 23 STRUCTURES | | Descriptor: | FACTOR VII | | Authors: | Muranyi, A, Finn, B.E, Gippert, G.P, Forsen, S, Stenflo, J, Drakenberg, T. | | Deposit date: | 1998-05-28 | | Release date: | 1999-02-16 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal EGF-like domain from human factor VII.
Biochemistry, 37, 1998
|
|
2V6E
 
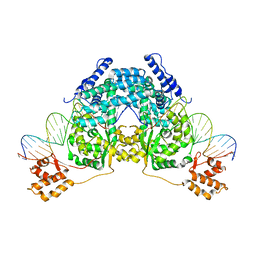 | |
2UUH
 
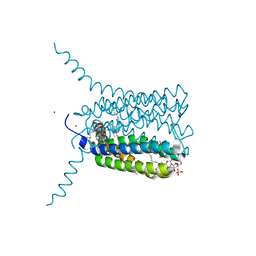 | | Crystal structure of Human Leukotriene C4 Synthase in complex with substrate glutathione | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, GLUTATHIONE, LEUKOTRIENE C4 SYNTHASE, ... | | Authors: | Martinez Molina, D, Wetterholm, A, Kohl, A, McCarthy, A.A, Niegowski, D, Ohlson, E, Hammarberg, T, Eshaghi, S, Haeggstrom, J.Z, Nordlund, P. | | Deposit date: | 2007-03-02 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Synthesis of Inflammatory Mediators by Human Leukotriene C4 Synthase.
Nature, 448, 2007
|
|
1BNK
 
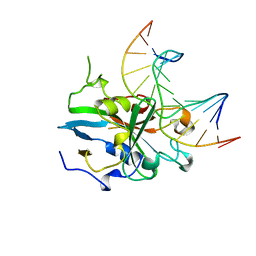 | | HUMAN 3-METHYLADENINE DNA GLYCOSYLASE COMPLEXED TO DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*YRRP*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), PROTEIN (3-METHYLADENINE DNA GLYCOSYLASE) | | Authors: | Lau, A.Y, Schaerer, O.D, Samson, L, Verdine, G.L, Ellenberger, T. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision.
Cell(Cambridge,Mass.), 95, 1998
|
|
1APO
 
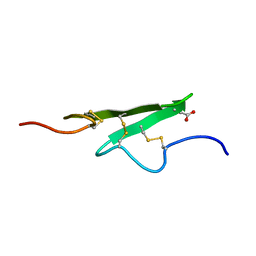 | | THREE-DIMENSIONAL STRUCTURE OF THE APO FORM OF THE N-TERMINAL EGF-LIKE MODULE OF BLOOD COAGULATION FACTOR X AS DETERMINED BY NMR SPECTROSCOPY AND SIMULATED FOLDING | | Descriptor: | EGF-LIKE MODULE OF BLOOD COAGULATION FACTOR X, HYDROXIDE ION | | Authors: | Ullner, M, Selander, M, Persson, E, Stenflo, J, Drakenberg, T, Teleman, O. | | Deposit date: | 1992-04-21 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the apo form of the N-terminal EGF-like module of blood coagulation factor X as determined by NMR spectroscopy and simulated folding.
Biochemistry, 31, 1992
|
|
7B4T
 
 | | Broadly neutralizing DARPin bnD.1 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.1, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), SULFATE ION | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4U
 
 | | Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.2, CALCIUM ION, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4V
 
 | | Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.2, CHLORIDE ION, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), ... | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4W
 
 | | Broadly neutralizing DARPin bnD.3 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Broadly neutralizing DARPin bnD.3, ... | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
2VEO
 
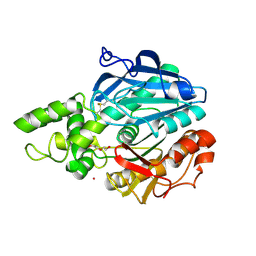 | | X-ray structure of Candida antarctica lipase A in its closed state. | | Descriptor: | GLYCEROL, LIPASE A, TETRAETHYLENE GLYCOL, ... | | Authors: | Ericsson, D.J, Kasrayan, A, Johansson, P, Bergfors, T, Sandstrom, A.G, Backvall, J.E, Mowbray, S.L. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure of Candida Antarctica Lipase a Shows a Novel Lid Structure and a Likely Mode of Interfacial Activation.
J.Mol.Biol., 376, 2008
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | Descriptor: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | Authors: | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | Deposit date: | 2005-09-24 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2PTL
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN-BINDING DOMAIN OF PROTEIN L. COMPARISON WITH THE IGG-BINDING DOMAINS OF PROTEIN G | | Descriptor: | PROTEIN L | | Authors: | Wikstroem, M, Drakenberg, T, Forsen, S, Sjoebring, U, Bjoerck, L. | | Deposit date: | 1994-08-12 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G.
Biochemistry, 33, 1994
|
|
1X9M
 
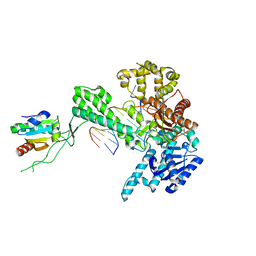 | | T7 DNA polymerase in complex with an N-2-acetylaminofluorene-adducted DNA | | Descriptor: | 5'-D(*CP*CP*CP*(8FG)P*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*TP*GP*GP*TP*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-23 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2IQC
 
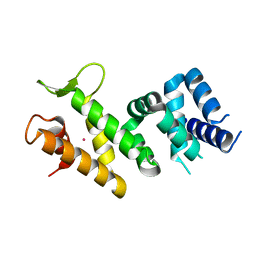 | | Crystal structure of Human FancF Protein that Functions in the Assembly of a DNA Damage Signaling Complex | | Descriptor: | Fanconi anemia group F protein, MERCURY (II) ION | | Authors: | Kowal, P, Gurtan, A.M, Stuckert, P, Lehmann, C, D'Andrea, A, Ellenberger, T.E. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex.
J.Biol.Chem., 282, 2007
|
|
1PU6
 
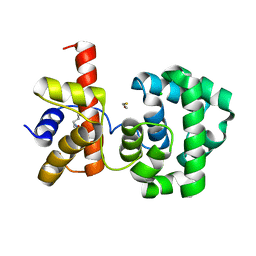 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-METHYLADENINE DNA GLYCOSYLASE, BETA-MERCAPTOETHANOL, ... | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PU7
 
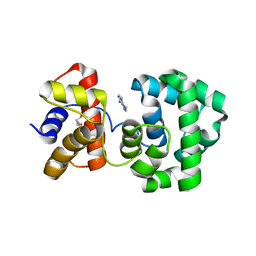 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 3,9-dimethyladenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 6-AMINO-3,9-DIMETHYL-9H-PURIN-3-IUM, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1NYH
 
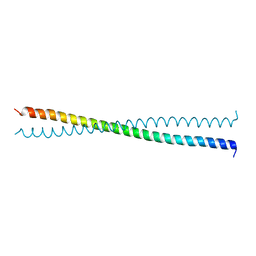 | | Crystal Structure of the Coiled-coil Dimerization Motif of Sir4 | | Descriptor: | Regulatory protein SIR4 | | Authors: | Chang, J.F, Hall, B.E, Tanny, J.C, Moazed, D, Filman, D, Ellenberger, T. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Coiled-coil Dimerization Motif of Sir4 and Its Interaction With Sir3
Structure, 11, 2003
|
|
1Q57
 
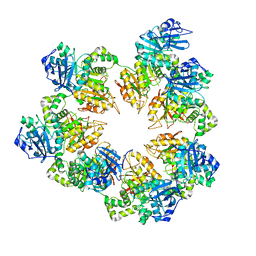 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
6TLB
 
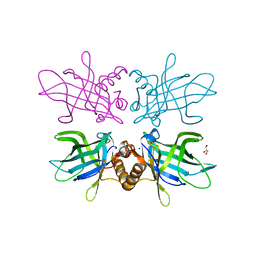 | | Plasmodium falciparum lipocalin (PF3D7_0925900) | | Descriptor: | GLYCEROL, SODIUM ION, Serine/threonine protein kinase | | Authors: | Burda, P.C, Crosskey, T.D, Lauk, K, Wilmanns, M, Gilberger, T.W. | | Deposit date: | 2019-12-02 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (2.85 Å), X-RAY DIFFRACTION | | Cite: | Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum.
Cell Rep, 31, 2020
|
|
1PU8
 
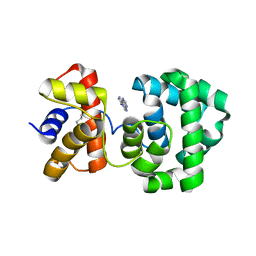 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 1,N6-ethenoadenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 3H-IMIDAZO[2,1-I]PURINE, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1CR1
 
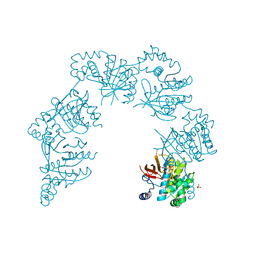 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTTP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR4
 
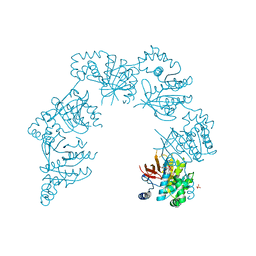 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTDP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
