3P0Q
 
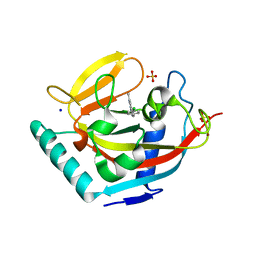 | | Human Tankyrase 2 - Catalytic PARP domain in complex with an inhibitor | | Descriptor: | N-[2-(4-chlorophenyl)ethyl]-6-methyl[1,2,4]triazolo[4,3-b]pyridazin-8-amine, SODIUM ION, SULFATE ION, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
3P0N
 
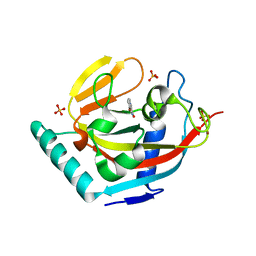 | | Human Tankyrase 2 - Catalytic PARP domain in complex with an inhibitor | | Descriptor: | 7-bromopyrrolo[1,2-a]quinoxalin-4(5H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
6Z5Q
 
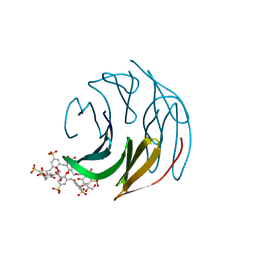 | | The RSL - sulfonato-calix[8]arene complex, P3 form, acetate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5P
 
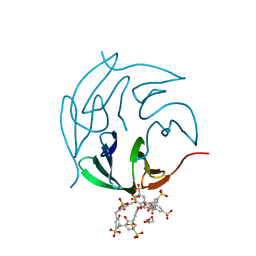 | | The RSL-R8 - sulfonato-calix[8]arene complex, P3 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SULFATE ION, ... | | Authors: | Ramberg, K, Skorek, T, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z62
 
 | | The RSL - sulfonato-calix[8]arene complex, P213 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.156 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5M
 
 | | The RSL - sulfonato-calix[8]arene complex, I23 form, Gly-HCl pH 2.2 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5Z
 
 | | The RSL-R6 - sulfonato-calix[8]arene complex, P213 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, beta-D-fructopyranose, sulfonato-calix[8]arene | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z60
 
 | | The RSL - sulfonato-calix[8]arene complex, P213 form, CAPS pH 9.5 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.166 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
1QQ9
 
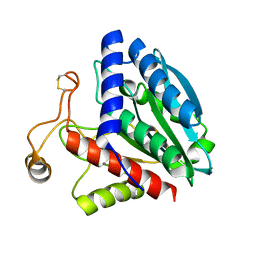 | | STREPTOMYCES GRISEUS AMINOPEPTIDASE COMPLEXED WITH METHIONINE | | Descriptor: | AMINOPEPTIDASE, CALCIUM ION, METHIONINE, ... | | Authors: | Gilboa, R, Greenblatt, H.M, Perach, M, Spungin-Bialik, A, Lessel, U, Schomburg, D, Blumberg, S, Shoham, G. | | Deposit date: | 1999-06-12 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Interactions of Streptomyces griseus aminopeptidase with a methionine product analogue: a structural study at 1.53 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4V6U
 
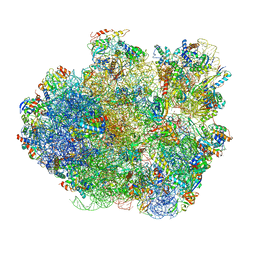 | | Promiscuous behavior of proteins in archaeal ribosomes revealed by cryo-EM: implications for evolution of eukaryotic ribosomes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10P, ... | | Authors: | Armache, J.-P, Anger, A.M, Marquez, V, Frankenberg, S, Froehlich, T, Villa, E, Berninghausen, O, Thomm, M, Arnold, G.J, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution.
Nucleic Acids Res., 41, 2013
|
|
2YF0
 
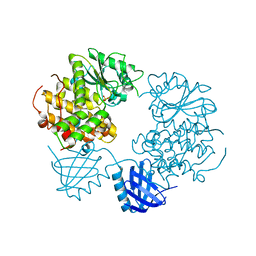 | | Human Myotubularin related protein 6 (MTMR6) | | Descriptor: | MYOTUBULARIN-RELATED PROTEIN 6, SULFATE ION | | Authors: | Moche, M, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Kouznetsova, E, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, VanDenBerg, S, Wahlberg, E, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Human Mtmr6
To be Published
|
|
2X8A
 
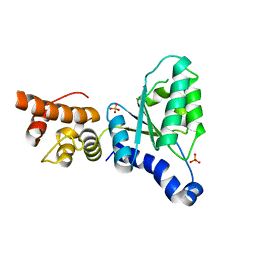 | | Human Nuclear Valosin containing protein Like (NVL), C-terminal AAA- ATPase domain | | Descriptor: | NUCLEAR VALOSIN-CONTAINING PROTEIN-LIKE, PHOSPHATE ION | | Authors: | Moche, M, Schuetz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Kraulis, P, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, VanDenBerg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H. | | Deposit date: | 2010-03-08 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Nuclear Valosin Containing Protein Like (Nvl) , C-Terminal Aaa-ATPase Domain
To be Published
|
|
3ZUI
 
 | | OMCI in complex with palmitoleic acid | | Descriptor: | COMPLEMENT INHIBITOR, PALMITOLEIC ACID | | Authors: | Roversi, P, Maillet, I, Togbe, D, Couillin, I, Quesniaux, V.F.J, Teixeira, M, Ahmat, N, Lissina, O, Boland, W, Ploss, K, Caesar, J.J.E, Leonhartsberger, S, Ryffel, B, Lea, S.M, Nunn, M.A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Bifunctional Lipocalin Ameliorates Murine Immune Complex-Induced Acute Lung Injury.
J.Biol.Chem., 288, 2013
|
|
3ZUO
 
 | | OMCI in complex with leukotriene B4 | | Descriptor: | COMPLEMENT INHIBITOR, LEUKOTRIENE B4 | | Authors: | Roversi, P, Maillet, I, Togbe, D, Couillin, I, Quesniaux, V.F.J, Teixeira, M, Ahmat, N, Lissina, O, Boland, W, Ploss, K, Caesar, J.J.E, Leonhartsberger, S, Ryffel, B, Lea, S.M, Nunn, M.A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Bifunctional Lipocalin Ameliorates Murine Immune Complex-Induced Acute Lung Injury.
J.Biol.Chem., 288, 2013
|
|
2WNS
 
 | | Human Orotate phosphoribosyltransferase (OPRTase) domain of Uridine 5' -monophosphate synthase (UMPS) in complex with its substrate orotidine 5'-monophosphate (OMP) | | Descriptor: | CHLORIDE ION, OROTATE PHOSPHORIBOSYLTRANSFERASE, OROTIDINE-5'-MONOPHOSPHATE | | Authors: | Moche, M, Roos, A, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotyenova, T, Kotzch, A, Nielsen, T.K, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, VanDenBerg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P. | | Deposit date: | 2009-07-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Orotate Phosphoribosyltransferase (Oprtase) Domain of Uridine 5'-Monophosphate Synthase (Umps) in Complex with its Substrate Orotidine 5'-Monophosphate (Omp)
To be Published
|
|
1GNS
 
 | | SUBTILISIN BPN' | | Descriptor: | ACETONE, SUBTILISIN BPN' | | Authors: | Almog, O, Gallagher, D.T, Ladner, J.E, Strausberg, S, Alexander, P. | | Deposit date: | 2001-10-06 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Thermostability. Analysis of Stabilizing Mutations in Subtilisin Bpn'.
J.Biol.Chem., 277, 2002
|
|
6EN6
 
 | | Crystal structure B of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
6EN5
 
 | | Crystal structure A of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6VQ2
 
 | | HLA-B*27:05 presenting an HIV-1 14mer peptide | | Descriptor: | 14-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQE
 
 | | HLA-B*27:05 presenting an HIV-1 13mer peptide | | Descriptor: | 13-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQZ
 
 | | HLA-B*27:05 presenting an HIV-1 6mer peptide | | Descriptor: | 6-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VPZ
 
 | | HLA-B*27:05 presenting an HIV-1 11mer peptide | | Descriptor: | 11-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQD
 
 | | HLA-B*27:05 presenting an HIV-1 8mer peptide | | Descriptor: | 8-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQY
 
 | | HLA-B*27:05 presenting an HIV-1 7mer peptide | | Descriptor: | 7-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
