1J8B
 
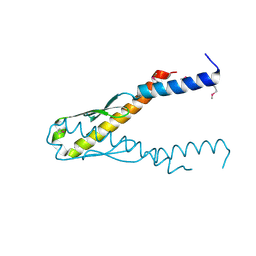 | | Structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function | | Descriptor: | YbaB | | Authors: | Lim, K, Tempcyzk, A, Toedt, J, Parsons, J.F, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-21 | | Release date: | 2003-01-14 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of YbaB from Haemophilus influenzae (HI0442), a
protein of unknown function coexpressed with the recombinational
DNA repair protein RecR
Proteins, 50, 2003
|
|
1IM8
 
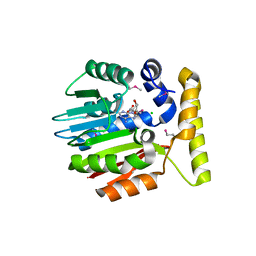 | | Crystal structure of YecO from Haemophilus influenzae (HI0319), a methyltransferase with a bound S-adenosylhomocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOSELENOCYSTEINE, YecO | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine.
Proteins, 45, 2001
|
|
4OJO
 
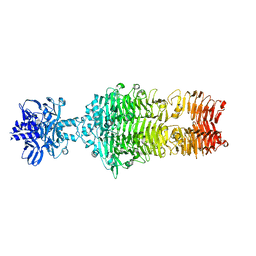 | |
4OJP
 
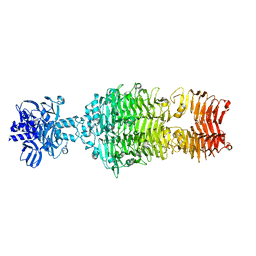 | |
1J8D
 
 | |
4OJL
 
 | |
1NU0
 
 | | Structure of the double mutant (L6M; F134M, SeMet form) of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF, SULFATE ION | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-30 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
2GPR
 
 | | GLUCOSE PERMEASE IIA FROM MYCOPLASMA CAPRICOLUM | | Descriptor: | GLUCOSE-PERMEASE IIA COMPONENT | | Authors: | Huang, K, Herzberg, O. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A promiscuous binding surface: crystal structure of the IIA domain of the glucose-specific permease from Mycoplasma capricolum.
Structure, 6, 1998
|
|
1R8G
 
 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
2HJP
 
 | |
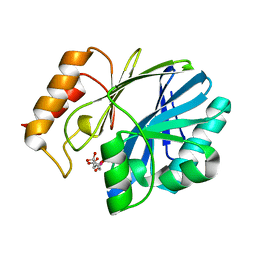
1MQO
 
 | |
1LVG
 
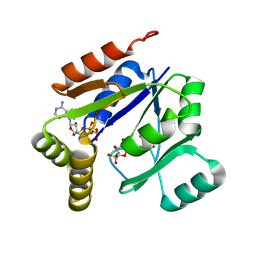 | | Crystal structure of mouse guanylate kinase in complex with GMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, ... | | Authors: | Sekulic, N, Shuvalova, L, Spangenberg, O, Konrad, M, Lavie, A. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the closed
conformation of mouse guanylate kinase.
J.Biol.Chem., 277, 2002
|
|
2HRW
 
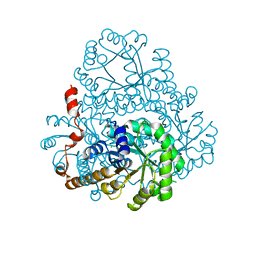 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
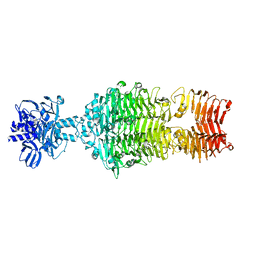
4OJ5
 
 | |
1NMN
 
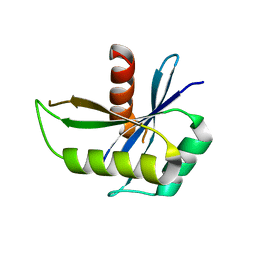 | | Structure of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
1SLT
 
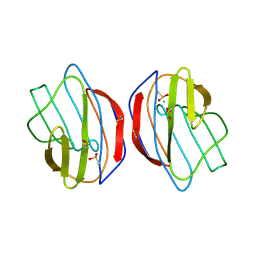 | |
1KO2
 
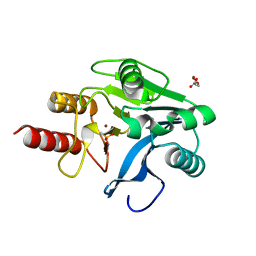 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with an oxidized Cys (cysteinesulfonic) | | Descriptor: | ACETATE ION, VIM-2 metallo-beta-lactamase, ZINC ION | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
1NNX
 
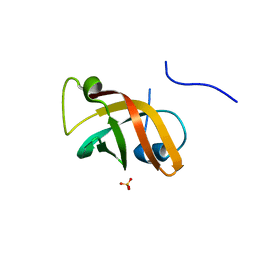 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
2UWX
 
 | | Active site restructuring regulates ligand recognition in class A penicillin-binding proteins | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, DiGuilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1MW5
 
 | | Structure of HI1480 from Haemophilus influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI1480 | | Authors: | Lim, K, Sarikaya, E, Howard, A, Galkin, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-27 | | Release date: | 2003-11-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1JO0
 
 | | Structure of HI1333, a Hypothetical Protein from Haemophilus influenzae with Structural Similarity to RNA-binding Proteins | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN HI1333 | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae
Proteins, 49, 2002
|
|
2FPH
 
 | |
1K25
 
 | |
1RW1
 
 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
2ISW
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
J.Biol.Chem., 282, 2007
|
|
