4XUZ
 
 | | Structure of CTX-M-15 bound to RPX-7009 at 1.5 A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Gardberg, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of a Cyclic Boronic Acid beta-Lactamase Inhibitor (RPX7009) with Utility vs Class A Serine Carbapenemases.
J.Med.Chem., 58, 2015
|
|
3G8A
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoHcy in space group P61 | | Descriptor: | Ribosomal RNA small subunit methyltransferase G, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3DMH
 
 | | T. Thermophilus 16S rRNA N2 G1207 methyltransferase (RsmC) in complex with AdoMet and Guanosine | | Descriptor: | GUANOSINE, Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Thermus thermophilus 16 S rRNA Methyltransferase RsmC in Complex with Cofactor and Substrate Guanosine.
J.Biol.Chem., 283, 2008
|
|
6CK4
 
 | | G96A mutant of the PRPP riboswitch from T. mathranii bound to ppGpp | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Reiss, C.W, Knappenberger, A.J, Strobel, S.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA.
Elife, 7, 2018
|
|
3D3D
 
 | | Bacteriophage lambda lysozyme complexed with a chitohexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme, SULFATE ION | | Authors: | Leung, A.K.W, Berghuis, A.M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the lytic transglycosylase from bacteriophage lambda in complex with hexa-N-acetylchitohexaose
Biochemistry, 40, 2001
|
|
3G88
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoMet in space group P61 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ribosomal RNA small subunit methyltransferase G, S-ADENOSYLMETHIONINE | | Authors: | Demirci, H, Belardinelli, R, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
2Z2N
 
 | |
4PJT
 
 | | Structure of PARP1 catalytic domain bound to inhibitor BMN 673 | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Aoyagi-Scharber, M, Gardberg, A.S, Arakaki, T.L. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the inhibition of poly(ADP-ribose) polymerases 1 and 2 by BMN 673, a potent inhibitor derived from dihydropyridophthalazinone.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1914
 
 | | SIGNAL RECOGNITION PARTICLE ALU RNA BINDING HETERODIMER, SRP9/14 | | Descriptor: | BETA-MERCAPTOETHANOL, PHOSPHATE ION, SIGNAL RECOGNITION PARTICLE 9/14 FUSION PROTEIN | | Authors: | Birse, D, Kapp, U, Strub, K, Cusack, S, Aberg, A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14.
EMBO J., 16, 1997
|
|
4Q02
 
 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | 3,4-difluorobenzenethiol, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
4Q03
 
 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | 4-bromobenzenethiol, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
2Z2O
 
 | |
2MMG
 
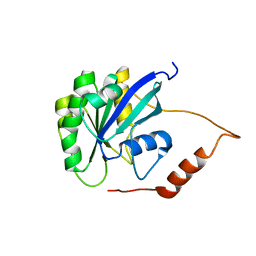 | | Structural Characterization of the Mengovirus Leader Protein Bound to Ran GTPase by Nuclear Magnetic Resonance | | Descriptor: | GTP-binding nuclear protein Ran | | Authors: | Bacot-Davis, V.R, Palmenberg, A.C, Cornilescu, C.C, Markley, J.L. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PZY
 
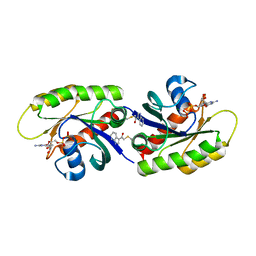 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | 2-chloro-1-(1H-indol-3-yl)ethanone, GUANOSINE-5'-DIPHOSPHATE, K-Ras, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
2NXC
 
 | |
4Q01
 
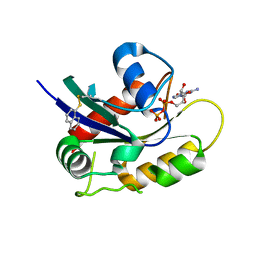 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras, MAGNESIUM ION, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
4PZZ
 
 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | 1H-benzimidazol-2-ylmethanethiol, GUANOSINE-5'-DIPHOSPHATE, K-Ras, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.403 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
2NXN
 
 | | T. thermophilus ribosomal protein L11 methyltransferase (PrmA) in complex with ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11, Ribosomal protein L11 methyltransferase | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA.
Embo J., 26, 2007
|
|
3CJQ
 
 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with dimethylated ribosomal protein L11 in space group P212121 | | Descriptor: | 50S ribosomal protein L11, IODIDE ION, N,N-dimethyl-L-methionine, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
4O9H
 
 | | Structure of Interleukin-6 in complex with a Camelid Fab fragment | | Descriptor: | Heavy Chain of the Camelid Fab fragment 61H7, Interleukin-6, Light Chain of the Camelid Fab fragment 61H7 | | Authors: | Klarenbeek, A, Blanchetot, C, Schragel, G, Sadi, A.S, Ongenae, N, Hemrika, W, Wijdenes, J, Spinelli, S, Desmyter, A, Cambillau, C, Hultberg, A, Kretz-rommel, A, Dreier, T, De haard, H.J.W, Roovers, R.C. | | Deposit date: | 2014-01-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Combining residues of naturally-occurring Camelid somatic affinity variants yields ultra-potent human therapeutic IL-6 antibodies
To be Published
|
|
2MMI
 
 | | Mengovirus Leader: Structural Characterization of the Mengovirus Leader Protein Bound to Ran GTPase by Nuclear Magnetic Resonance | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Cornilescu, C.C, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MML
 
 | | T47 phosphorylation of the Mengovirus Leader Protein: NMR Studies of the Phosphorylation of the Mengovirus Leader Protein Reveal Stabilization of Intermolecular Domain Interactions | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Porter, F.W, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CJT
 
 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with dimethylated ribosomal protein L11 | | Descriptor: | 1,2-ETHANEDIOL, 50S ribosomal protein L11, CHLORIDE ION, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
3CJS
 
 | | Minimal Recognition Complex between PrmA and Ribosomal Protein L11 | | Descriptor: | 1,2-ETHANEDIOL, 50S ribosomal protein L11, Ribosomal protein L11 methyltransferase | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
3CJR
 
 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with ribosomal protein L11 (K39A) and inhibitor Sinefungin. | | Descriptor: | 50S ribosomal protein L11, Ribosomal protein L11 methyltransferase, SINEFUNGIN | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
