1Q7G
 
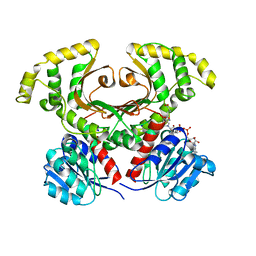 | | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline | | Descriptor: | Homoserine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE-5-HYDROXY-4-OXONORVALINE, SODIUM ION | | Authors: | Jacques, S.L, Mirza, I.A, Ejim, L, Koteva, K, Hughes, D.W, Green, K, Kinach, R, Honek, J.F, Lai, H.K, Berghuis, A.M, Wright, G.D. | | Deposit date: | 2003-08-18 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme assisted suicide: Molecular basis for the antifungal activity of 5-hydroxy-4-oxonorvaline by potent inhibition of homoserine dehydrogenase
Chem.Biol., 10, 2003
|
|
6YW5
 
 | | The structure of the small subunit of the mitoribosome from Neurospora crassa | | Descriptor: | 16S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
1QY2
 
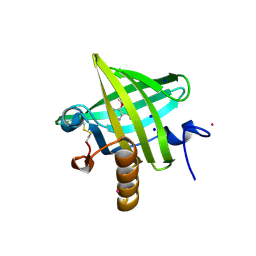 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOPROPYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
1BXE
 
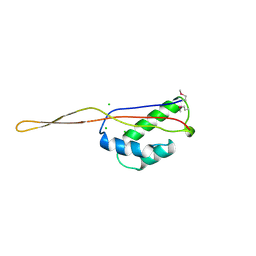 | | RIBOSOMAL PROTEIN L22 FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, PROTEIN (RIBOSOMAL PROTEIN L22) | | Authors: | Unge, J, Aberg, A, Al-Karadaghi, S, Nikulin, A, Nikonov, S, Davydova, N, Nevskaya, N, Garber, M, Liljas, A. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance.
Structure, 6, 1998
|
|
1QY1
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
2MMI
 
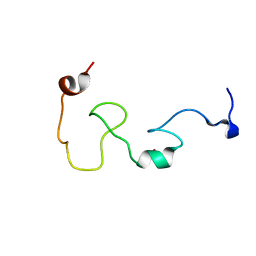 | | Mengovirus Leader: Structural Characterization of the Mengovirus Leader Protein Bound to Ran GTPase by Nuclear Magnetic Resonance | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Cornilescu, C.C, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MML
 
 | | T47 phosphorylation of the Mengovirus Leader Protein: NMR Studies of the Phosphorylation of the Mengovirus Leader Protein Reveal Stabilization of Intermolecular Domain Interactions | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Porter, F.W, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2NXJ
 
 | |
1A8P
 
 | | FERREDOXIN REDUCTASE FROM AZOTOBACTER VINELANDII | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:FERREDOXIN OXIDOREDUCTASE | | Authors: | Prasad, G.S, Kresge, N, Muhlberg, A.B, Shaw, A, Jung, Y.S, Burgess, B.K, Stout, C.D. | | Deposit date: | 1998-03-28 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of NADPH:ferredoxin reductase from Azotobacter vinelandii.
Protein Sci., 7, 1998
|
|
1A3K
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN GALECTIN-3 CARBOHYDRATE RECOGNITION DOMAIN (CRD) AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seetharaman, J, Kanigsberg, A, Slaaby, R, Leffler, H, Barondes, S.H, Rini, J.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution.
J.Biol.Chem., 273, 1998
|
|
2CM0
 
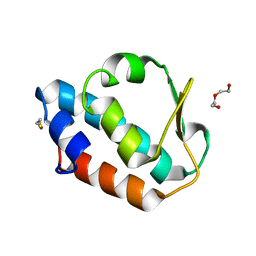 | |
1GIL
 
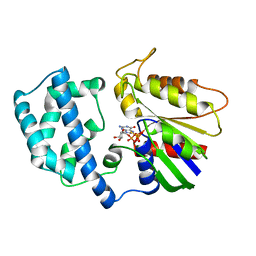 | |
1QY0
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | CADMIUM ION, GLYCEROL, Major urinary protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
1ADY
 
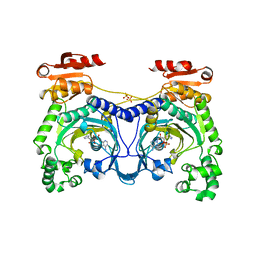 | | HISTIDYL-TRNA SYNTHETASE IN COMPLEX WITH HISTIDYL-ADENYLATE | | Descriptor: | HISTIDYL-ADENOSINE MONOPHOSPHATE, HISTIDYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Cusack, S, Aberg, A. | | Deposit date: | 1997-02-19 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase.
Biochemistry, 36, 1997
|
|
2B0Q
 
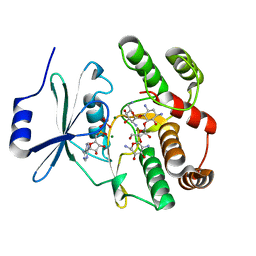 | |
2GN9
 
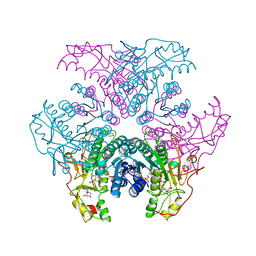 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-Glc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2NXE
 
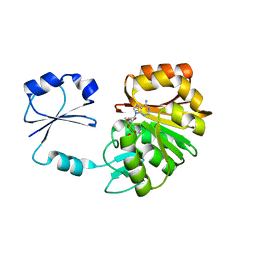 | | T. thermophilus ribosomal protein L11 methyltransferase (PrmA) in complex with S-Adenosyl-L-Methionine | | Descriptor: | Ribosomal protein L11 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA.
Embo J., 26, 2007
|
|
2GN6
 
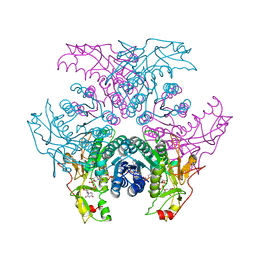 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-GlcNAc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
1GFI
 
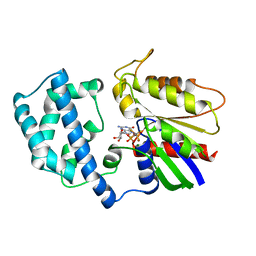 | | STRUCTURES OF ACTIVE CONFORMATIONS OF GI ALPHA 1 AND THE MECHANISM OF GTP HYDROLYSIS | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Coleman, D.E, Berghuis, A.M, Sprang, S.R. | | Deposit date: | 1994-11-11 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis.
Science, 265, 1994
|
|
1ADJ
 
 | | HISTIDYL-TRNA SYNTHETASE IN COMPLEX WITH HISTIDINE | | Descriptor: | HISTIDINE, HISTIDYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Cusack, S, Aberg, A. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase.
Biochemistry, 36, 1997
|
|
2MMH
 
 | | Unphosphorylated Mengovirus Leader Protein: NMR Studies of the Phosphorylation of the Mengovirus Leader Protein Reveal Stabilization of Intermolecular Domain Interactions | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Porter, F.W, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2GN8
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
1EBU
 
 | | HOMOSERINE DEHYDROGENASE COMPLEX WITH NAD ANALOGUE AND L-HOMOSERINE | | Descriptor: | 3-AMINOMETHYL-PYRIDINIUM-ADENINE-DINUCLEOTIDE, HOMOSERINE DEHYDROGENASE, L-HOMOSERINE, ... | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
2GNA
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-Gal | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GALACTOSE-URIDINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2B7L
 
 | | Crystal Structure of CTP:Glycerol-3-Phosphate Cytidylyltransferase from Staphylococcus aureus | | Descriptor: | glycerol-3-phosphate cytidylyltransferase | | Authors: | Fong, D.H, Yim, V.C.-N, D'Elia, M.A, Brown, E.D, Berghuis, A.M. | | Deposit date: | 2005-10-04 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of CTP:glycerol-3-phosphate cytidylyltransferase from Staphylococcus aureus: examination of structural basis for kinetic mechanism.
Biochim.Biophys.Acta, 1764, 2006
|
|
