5LQ4
 
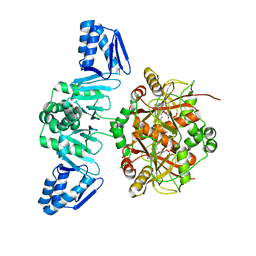 | | The Structure of ThcOx, the First Oxidase Protein from the Cyanobactin Pathways | | Descriptor: | CyaGox, FLAVIN MONONUCLEOTIDE | | Authors: | Bent, A.F, Wagner, A, Naismith, J.H. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the cyanobactin oxidase ThcOx from Cyanothece sp. PCC 7425, the first structure to be solved at Diamond Light Source beamline I23 by means of S-SAD.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4BG2
 
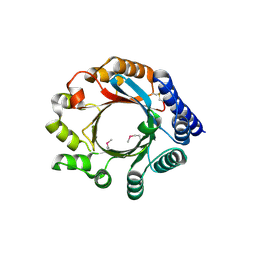 | | X-ray Crystal Structure of PatF from Prochloron didemni | | Descriptor: | PATF | | Authors: | Bent, A.F, Koehnke, J, Houssen, W.E, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of Patf from Prochloron Didemni.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5TEB
 
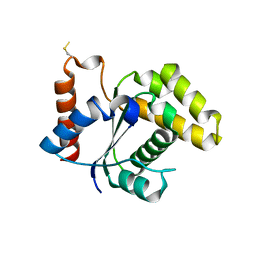 | | Crystal Structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPP1 | | Descriptor: | Recognition of Peronospora parasitica 1 | | Authors: | Bentham, A.R, Zhang, X, Croll, T, Williams, S, Kobe, B. | | Deposit date: | 2016-09-20 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8PQ7
 
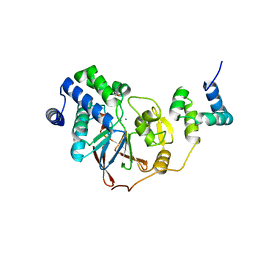 | |
8B2R
 
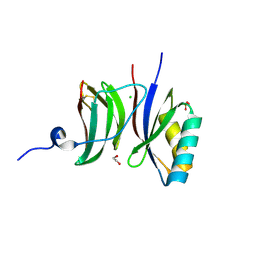 | |
7NLJ
 
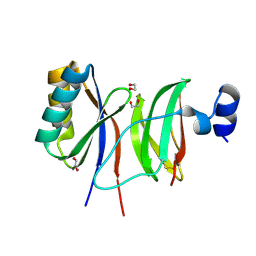 | |
7NMM
 
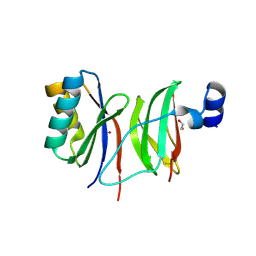 | |
4UVQ
 
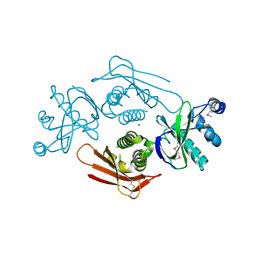 | | PatG Domain of Unknown Function | | Descriptor: | THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE, ZINC ION | | Authors: | Mann, G, Koehnke, J, Bent, A.F, Graham, R, Schwarz-Linek, U, Naismith, J.H. | | Deposit date: | 2014-08-07 | | Release date: | 2014-09-17 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | The Structure of the Cyanobactin Domain of Unknown Function from Patg in the Patellamide Gene Cluster
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5O3U
 
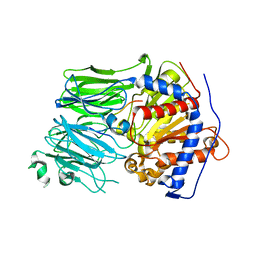 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin F1 | | Descriptor: | Peptide cyclase 1, Putative presegetalin F1 | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5NJN
 
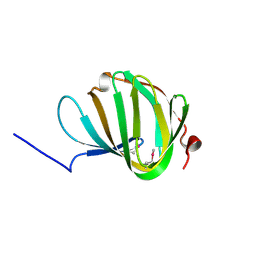 | | Roll out the beta-barrel: structure and mechanism of Pac13, a unique nucleoside dehydratase | | Descriptor: | Putative cupin_2 domain-containing isomerase | | Authors: | Michailidou, F, Bent, A.F, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2017-03-29 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NJO
 
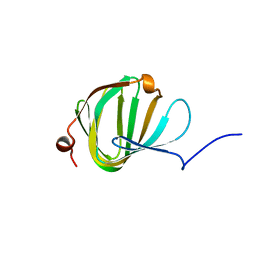 | | Roll out the beta-barrel: structure and mechanism of Pac13, a unique nucleoside dehydratase | | Descriptor: | Putative cupin_2 domain-containing isomerase | | Authors: | Michailidou, F, Bent, A.F, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2017-03-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O3X
 
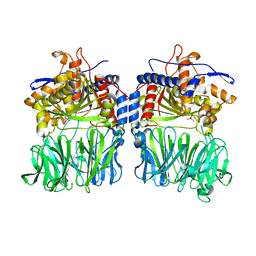 | | Structural characterization of the fast and promiscuous macrocyclase from plant - apo PCY1 | | Descriptor: | CACODYLATE ION, Peptide cyclase 1 | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5O3V
 
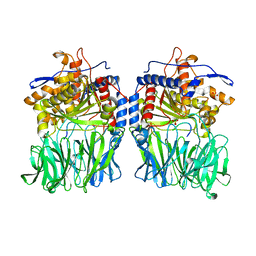 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin B1 | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Putative presegetalin B1, ... | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5O3W
 
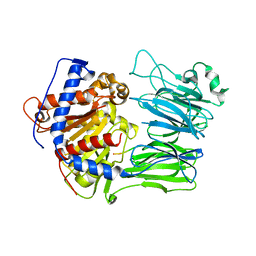 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin A1 | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Presegetalin A1, ... | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
4BS9
 
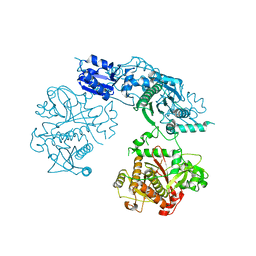 | | Structure of the heterocyclase TruD | | Descriptor: | TRUD, ZINC ION | | Authors: | Koehnke, J, Bent, A.F, Zollman, D, Smith, K, Houssen, W.E, Zhu, X, Mann, G, Lebl, T, Scharff, R, Shirran, S, Botting, C.H, Jaspars, M, Schwarz-Linek, U, Naismith, J.H. | | Deposit date: | 2013-06-09 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Cyanobactin Heterocyclase Enzyme: A Processive Adenylase that Operates with a Defined Order of Reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4AKT
 
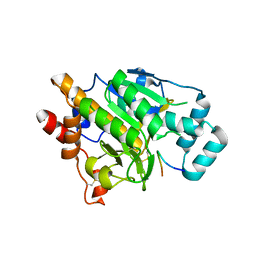 | | PatG macrocyclase in complex with peptide | | Descriptor: | SUBSTRATE ANALOGUE, THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AKS
 
 | | PatG macrocyclase domain | | Descriptor: | THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2WEG
 
 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | Descriptor: | 2-fluorobenzenesulfonamide, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | Deposit date: | 2009-03-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
2WEO
 
 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | Descriptor: | 3-fluorobenzenesulfonamide, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | Deposit date: | 2009-04-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
2WEJ
 
 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, ZINC ION, ... | | Authors: | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | Deposit date: | 2009-03-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
2WEH
 
 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | Descriptor: | 2-CHLOROBENZENESULFONAMIDE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | Deposit date: | 2009-03-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
2YJD
 
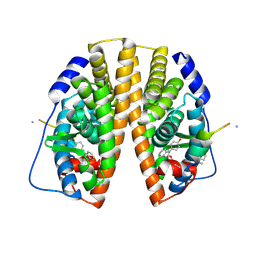 | | Stapled peptide bound to Estrogen Receptor Beta | | Descriptor: | 4-(2-PROPAN-2-YLOXYBENZIMIDAZOL-1-YL)PHENOL, ESTROGEN RECEPTOR BETA, STAPLED PEPTIDE | | Authors: | Phillips, C, Roberts, L.R, Schade, M, Bent, A, Davies, N.L, Moore, R, Pannifer, A.D, Brown, D.G, Pickford, A.R, Irving, S.L. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Structure of Stapled Peptides Binding to Estrogen Receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LDD
 
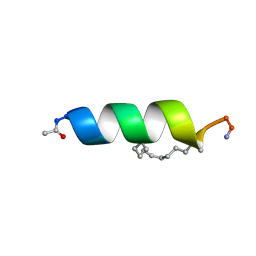 | | Solution structure of the estrogen receptor-binding stapled peptide SP6 (Ac-EKHKILXRLLXDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP6 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LDA
 
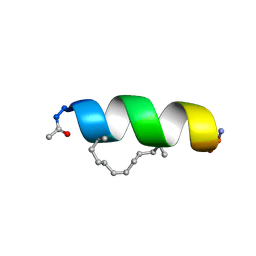 | | Solution structure of the estrogen receptor-binding stapled peptide SP2 (Ac-HKXLHQXLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP2 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LDC
 
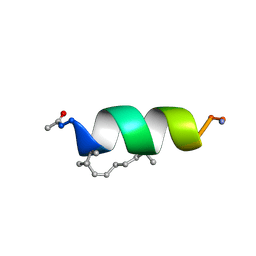 | | Solution structure of the estrogen receptor-binding stapled peptide SP1 (Ac-HXILHXLLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP1 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
