1LYJ
 
 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
8D4Z
 
 | | Crystal structure of USP7 in complex with allosteric inhibitor FX1-3763 | | Descriptor: | 1-({(7M)-7-[1-(azetidin-3-yl)-6-chloro-1,2,3,4-tetrahydroquinolin-8-yl]thieno[3,2-b]pyridin-2-yl}methyl)pyrrolidine-2,5-dione, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bell, J.A. | | Deposit date: | 2022-06-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel USP7 inhibitors demonstrate potent anti-cancer activity in models of AML, synergy with BCL2 inhibition, and a differentiated mechanism of action
To Be Published
|
|
9MJM
 
 | | SOS1 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-({(1R)-1-[2-methyl-3-(trifluoromethyl)phenyl]ethyl}amino)-3-(2-oxaspiro[3.3]heptan-6-yl)-5,6,7,8-tetrahydropyrido[4,3-d]pyrimidin-4(3H)-one, IMIDAZOLE, ... | | Authors: | Bell, J.A. | | Deposit date: | 2024-12-16 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploiting Solvent Exposed Salt-Bridge Interactions for the Discovery of Potent Inhibitors of SOS1 Using Free-Energy Perturbation Simulations.
Acs Med.Chem.Lett., 16, 2025
|
|
9MJL
 
 | | SOS1 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 6-({(1R)-1-[2-methyl-3-(trifluoromethyl)phenyl]ethyl}amino)-8-(oxan-4-yl)-1,3,4,8-tetrahydropyrido[3,4-c][1,6]naphthyridin-9(2H)-one, Son of sevenless homolog 1 | | Authors: | Bell, J.A. | | Deposit date: | 2024-12-16 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Exploiting Solvent Exposed Salt-Bridge Interactions for the Discovery of Potent Inhibitors of SOS1 Using Free-Energy Perturbation Simulations.
Acs Med.Chem.Lett., 16, 2025
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
8VSG
 
 | | SARS-CoV-2 main protease with covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-(1-phenylcyclopropane-1-carbonyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bell, J.A, Bandera, A.M. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
9D3V
 
 | | CRYSTAL STRUCTURE OF EGFR(L858R/T790M/C797S) IN COMPLEX WITH AUR-3418 | | Descriptor: | (4S)-N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-2-methyl-1-(propan-2-yl)-1H-imidazo[1,2-b]pyrazol-6-amine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Bell, J.A. | | Deposit date: | 2024-08-12 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of a Novel Mutant-Selective Epidermal Growth Factor Receptor Inhibitor Using an In Silico Enabled Drug Discovery Platform.
J.Med.Chem., 67, 2024
|
|
9D3W
 
 | | CRYSTAL STRUCTURE OF EGFR(L858R/T790M/C797S) IN COMPLEX WITH AUR-8250 | | Descriptor: | (4S)-1-(1,4-dioxaspiro[4.5]decan-8-yl)-N-{2-[(3S,4R)-3-fluoro-4-methoxypiperidin-1-yl]pyrimidin-4-yl}-2-methyl-1H-imidazo[1,2-b]pyrazol-6-amine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Bell, J.A. | | Deposit date: | 2024-08-12 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of a Novel Mutant-Selective Epidermal Growth Factor Receptor Inhibitor Using an In Silico Enabled Drug Discovery Platform.
J.Med.Chem., 67, 2024
|
|
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
1ORF
 
 | | The Oligomeric Structure of Human Granzyme A Reveals the Molecular Determinants of Substrate Specificity | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Granzyme A, SULFATE ION | | Authors: | Bell, J.K, Goetz, D.H, Mahrus, S, Harris, J.L, Fletterick, R.J, Craik, C.S. | | Deposit date: | 2003-03-12 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The oligomeric structure of human granzyme A is a determinant of its extended substrate specificity.
Nat.Struct.Biol., 10, 2003
|
|
4M9T
 
 | |
2A0Z
 
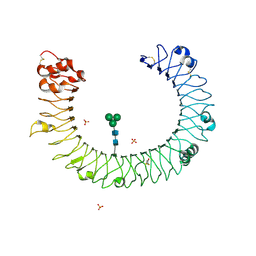 | | The molecular structure of toll-like receptor 3 ligand binding domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bell, J.K, Botos, I, Hall, P.R, Askins, J, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the Toll-like receptor 3 ligand-binding domain
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1SC6
 
 | |
1C0B
 
 | |
1C0C
 
 | |
1IE3
 
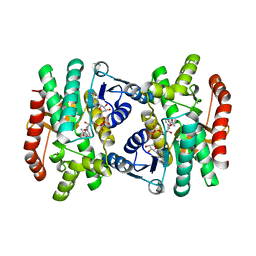 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1L26
 
 | |
1L30
 
 | |
1L28
 
 | |
1L32
 
 | |
1L25
 
 | |
1L31
 
 | |
1L29
 
 | |
