7OID
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 5A | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
7OI7
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 2 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
7OIA
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 3C | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
7OIC
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 4 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
7OIE
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 5B | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
7OIB
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 3D | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
3J7Z
 
 | | Structure of the E. coli 50S subunit with ErmCL nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Arenz, S, Meydan, S, Starosta, A.L, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Wilson, D.N. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Drug Sensing by the Ribosome Induces Translational Arrest via Active Site Perturbation.
Mol.Cell, 56, 2014
|
|
3J9Y
 
 | | Cryo-EM structure of tetracycline resistance protein TetM bound to a translating E.coli ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Nguyen, F, Beckmann, R, Wilson, D.N. | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-15 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9- angstrom resolution.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3J9W
 
 | | Cryo-EM structure of the Bacillus subtilis MifM-stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein bS16, ... | | Authors: | Sohmen, D, Chiba, S, Shimokawa-Chiba, N, Innis, C.A, Berninghausen, O, Beckmann, R, Ito, K, Wilson, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling.
Nat Commun, 6, 2015
|
|
6EM5
 
 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6G5I
 
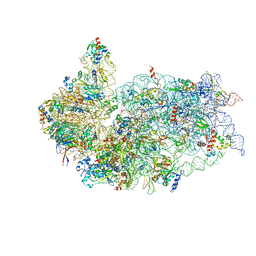 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State R | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G53
 
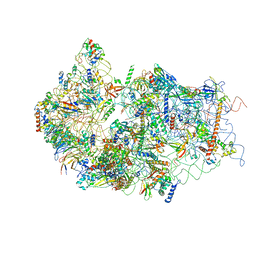 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State E | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G4S
 
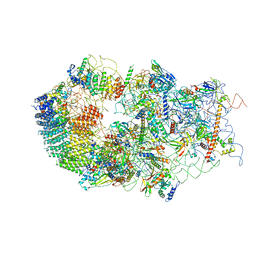 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State B | | Descriptor: | 40S ribosomal protein S11, 40S ribosomal protein S13, 40S ribosomal protein S14, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G5H
 
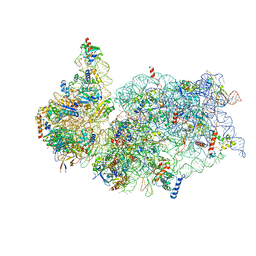 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - Mature | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G4W
 
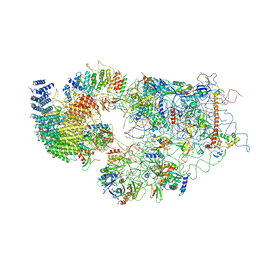 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State A | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G51
 
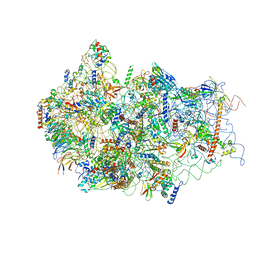 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State D | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
6HD7
 
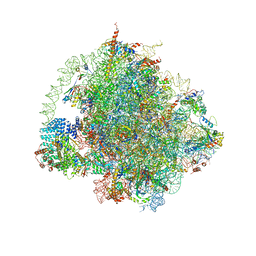 | | Cryo-EM structure of the ribosome-NatA complex | | Descriptor: | 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Becker, T, Beckmann, R. | | Deposit date: | 2018-08-17 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
3IZQ
 
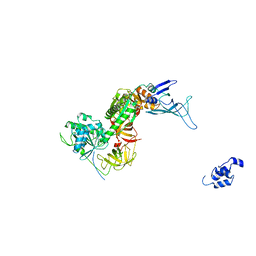 | | Structure of the Dom34-Hbs1-GDPNP complex bound to a translating ribosome | | Descriptor: | Elongation factor 1 alpha-like protein, Protein DOM34 | | Authors: | Becker, T, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Sieber, H, Abdel Motaal, B, Mielke, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2010-11-30 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3J2I
 
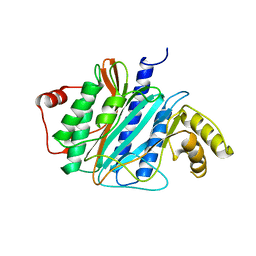 | | Structure of late pre-60S ribosomal subunits with nuclear export factor Arx1 bound at the peptide exit tunnel | | Descriptor: | Eukaryotic translation initiation factor 6, Proliferation-associated protein 2G4 | | Authors: | Bradatsch, B, Leidig, C, Granneman, S, Gnaedig, M, Tollervey, D, Boettcher, B, Beckmann, R, Hurt, E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3J25
 
 | | Structural basis for TetM-mediated tetracycline resistance | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tetracycline resistance protein tetM | | Authors: | Doenhoefer, A, Franckenberg, S, Wickles, S, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis for TetM-mediated tetracycline resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7Z20
 
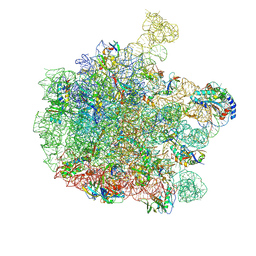 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
8K2D
 
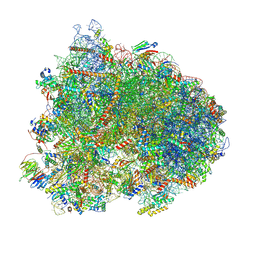 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, eEF2, Stm1 and eIF5A | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K82
 
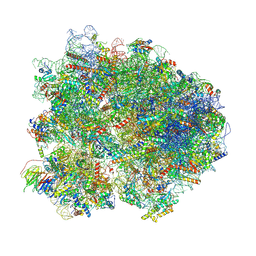 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, Not5 and P-site tRNA | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
6ZON
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
