8PV5
 
 | | Chaetomium thermophilum pre-60S State 8 - pre-5S rotation without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV4
 
 | | Chaetomium thermophilum pre-60S State 2 - pre-5S rotation with Rix1 complex - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV6
 
 | | Chaetomium thermophilum pre-60S State 3 - post-5S rotation with Rix1 complex with Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV1
 
 | | Chaetomium thermophilum pre-60S State 6 - pre-5S rotation - L1 intermediate - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVL
 
 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV8
 
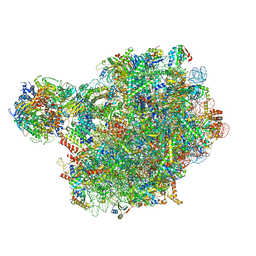 | | Chaetomium thermophilum pre-60S State 4 - post-5S rotation with Rix1 complex without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
5ABB
 
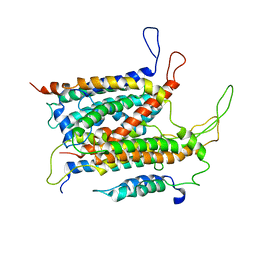 | | Visualization of a polytopic membrane protein during SecY-mediated membrane insertion | | Descriptor: | GREEN-LIGHT ABSORBING PROTEORHODOPSIN, PROTEIN TRANSLOCASE SUBUNIT SECE, PROTEIN TRANSLOCASE SUBUNIT SECY | | Authors: | Bischoff, L, Wickles, S, Berninghausen, O, vanderSluis, E, Beckmann, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Visualization of a Polytopic Membrane Protein During Secy-Mediated Membrane Insertion.
Nat.Commun., 5, 2014
|
|
6R86
 
 | | Yeast Vms1-60S ribosomal subunit complex (post-state) | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
6R87
 
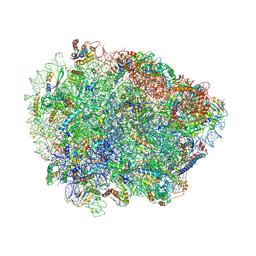 | | Yeast Vms1 (Q295L)-60S ribosomal subunit complex (pre-state without Arb1) | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
6R5Q
 
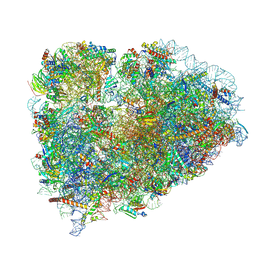 | | Structure of XBP1u-paused ribosome nascent chain complex (post-state) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shanmuganathan, V, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-07-10 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
7AJU
 
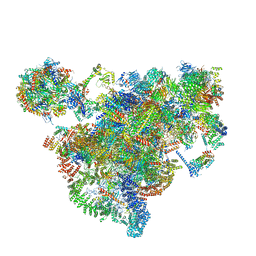 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7AJT
 
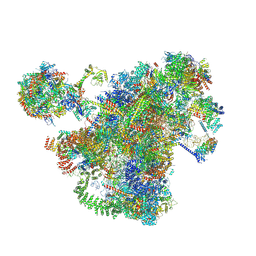 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
6SH3
 
 | |
6R6P
 
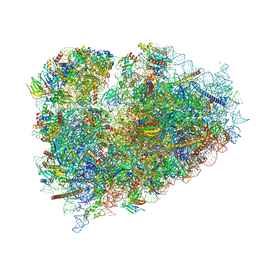 | | Structure of XBP1u-paused ribosome nascent chain complex (rotated state) | | Descriptor: | 18S rRNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shanmuganathan, V, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
6R7L
 
 | | Ribosome-bound SecYEG translocon in a nanodisc | | Descriptor: | Protein translocase subunit SecY, SecE,Protein translocase subunit SecE,Protein translocase subunit SecE,Protein translocase subunit SecE, SecG | | Authors: | Kater, L, Beckmann, R, Kedrov, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Partially inserted nascent chain unzips the lateral gate of the Sec translocon.
Embo Rep., 20, 2019
|
|
6R7Q
 
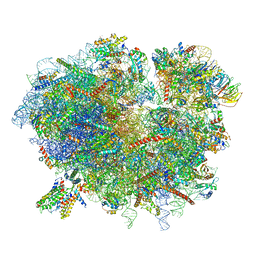 | | Structure of XBP1u-paused ribosome nascent chain complex with Sec61. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shanmuganathan, V, Cheng, J, Braunger, K, Berninghausen, O, Beatrix, B, Beckmann, R. | | Deposit date: | 2019-03-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
6R84
 
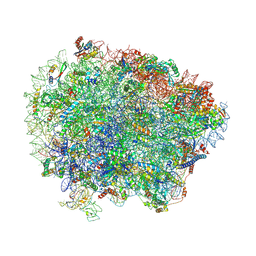 | | Yeast Vms1 (Q295L)-60S ribosomal subunit complex (pre-state with Arb1) | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
6ZQF
 
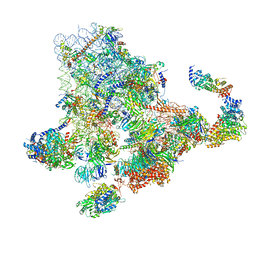 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-B (Poly-Ala) | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
1G61
 
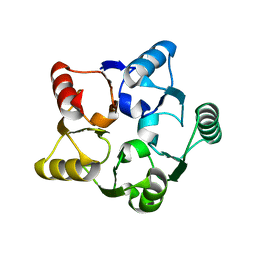 | | CRYSTAL STRUCTURE OF M.JANNASCHII EIF6 | | Descriptor: | TRANSLATION INITIATION FACTOR 6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
6ZXE
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State F2 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZXD
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State F1 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZXH
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State H2 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZZZ
 
 | |
6ZQB
 
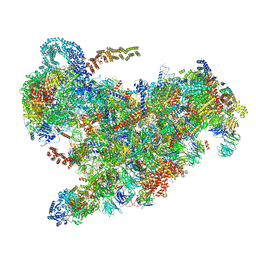 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state B2 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
