8FSD
 
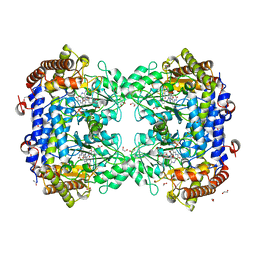 | | P130R mutant of soybean SHMT8 in complex with PLP-glycine and formylTHF | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Beamer, L.J, Korasick, D.A. | | Deposit date: | 2023-01-09 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional analysis of two SHMT8 variants associated with soybean cyst nematode resistance.
Febs J., 291, 2024
|
|
9CE6
 
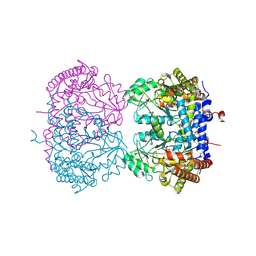 | |
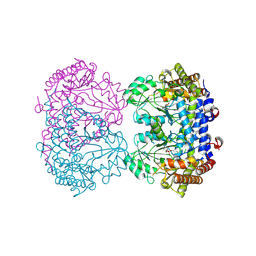
9CG8
 
 | |
5JN5
 
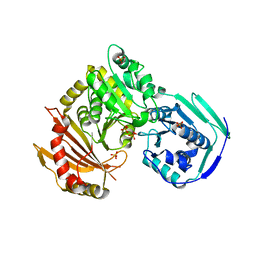 | |
5EPC
 
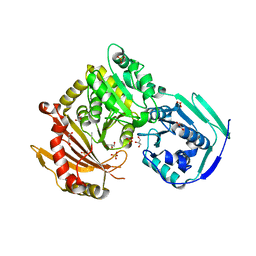 | | Crystal structure of wild-type human phosphoglucomutase 1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphoglucomutase-1, ... | | Authors: | Beamer, L.J. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Induced Structural Disorder as a Molecular Mechanism for Enzyme Dysfunction in Phosphoglucomutase 1 Deficiency.
J.Mol.Biol., 428, 2016
|
|
5F9C
 
 | |
1LMB
 
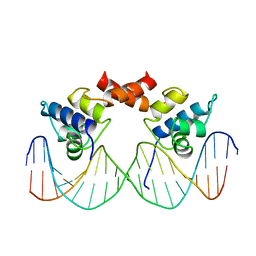 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
3OLP
 
 | |
1BP1
 
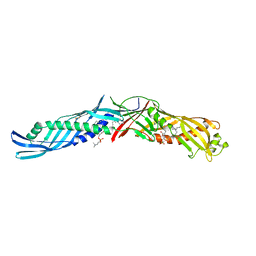 | | CRYSTAL STRUCTURE OF BPI, THE HUMAN BACTERICIDAL PERMEABILITY-INCREASING PROTEIN | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Beamer, L.J, Carroll, S.F, Eisenberg, D. | | Deposit date: | 1997-04-08 | | Release date: | 1997-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution.
Science, 276, 1997
|
|
6UO6
 
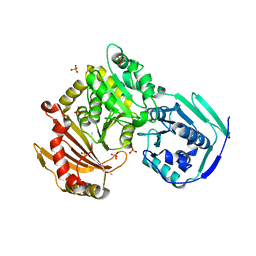 | |
2H4L
 
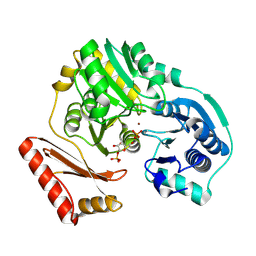 | | Complex of PMM/PGM with ribose 1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Beamer, L.J. | | Deposit date: | 2006-05-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complexes of the enzyme phosphomannomutase/phosphoglucomutase with a slow substrate and an inhibitor.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5TR2
 
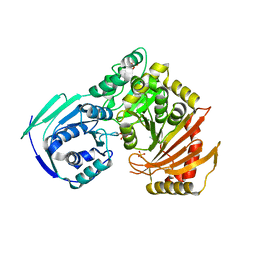 | |
6MNV
 
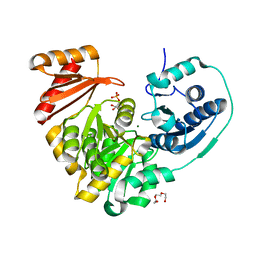 | | Crystal structure of X. citri phosphoglucomutase in complex with CH2FG1P | | Descriptor: | 1-deoxy-1-fluoro-2-O-phosphono-alpha-D-gluco-hept-2-ulopyranose, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase, ... | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-10-03 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
6MLF
 
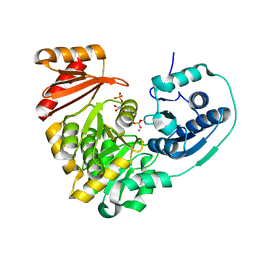 | |
6MLW
 
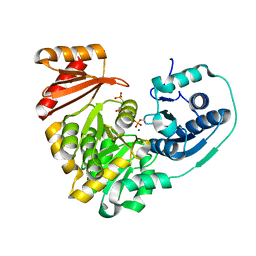 | | Crystal structure of X. citri phosphoglucomutase in complex with 2-fluoro mannosyl-1-methyl-phosphonic acid | | Descriptor: | 2,6-anhydro-5,7-dideoxy-5-fluoro-7-phosphono-D-glycero-D-manno-heptitol, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-09-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
6MLH
 
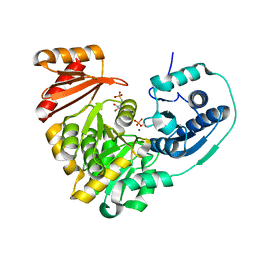 | | Crystal structure of X. citri phosphoglucomutase in complex with GLUCOPYRANOSYL-1-METHYL-PHOSPHONIC ACID | | Descriptor: | (1S)-1,5-anhydro-1-(phosphonomethyl)-D-glucitol, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-09-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
6N1E
 
 | | Crystal structure of X. citri phosphoglucomutase in complex with 1-methyl-glucose 6-phosphate | | Descriptor: | 1-deoxy-7-O-phosphono-alpha-D-gluco-hept-2-ulopyranose, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Beamer, L.J, Stiers, K.M. | | Deposit date: | 2018-11-08 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis, Derivatization, and Structural Analysis of Phosphorylated Mono-, Di-, and Trifluorinated d-Gluco-heptuloses by Glucokinase: Tunable Phosphoglucomutase Inhibition.
Acs Omega, 4, 2019
|
|
1U6D
 
 | |
1EWF
 
 | | THE 1.7 ANGSTROM CRYSTAL STRUCTURE OF BPI | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Kleiger, G, Beamer, L.J, Grothe, R, Mallick, P, Eisenberg, D. | | Deposit date: | 2000-04-25 | | Release date: | 2000-06-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of BPI: a study of how two dissimilar amino acid sequences can adopt the same fold.
J.Mol.Biol., 299, 2000
|
|
8DOM
 
 | |
8TQF
 
 | | Crystal structure of Soybean SHMT8 in complex with PLP-glycine and diglutamylated 5-formyltetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6R)-2-amino-5-formyl-4-hydroxy-5,6,7,8-tetrahydropteridin-6-yl]methyl}amino)benzoyl]-L-gamma-glutamyl-L-glutamic acid, ... | | Authors: | Owuocha, L.F, Beamer, L.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural insights into high affinity binding of polyglutamylated tetrahydrofolate to soybean serine hydroxymethyltransferase
To Be Published
|
|
3NA5
 
 | | Crystal structure of a bacterial phosphoglucomutase, an enzyme important in the virulence of several human pathogens. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Mehra-Chaudhary, R, Beamer, L.J. | | Deposit date: | 2010-06-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens.
Proteins, 79, 2011
|
|
4MRQ
 
 | | Crystal Structure of wild-type unphosphorylated PMM/PGM | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, ... | | Authors: | Lee, Y, Beamer, L. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Promotion of enzyme flexibility by dephosphorylation and coupling to the catalytic mechanism of a phosphohexomutase.
J.Biol.Chem., 289, 2014
|
|
1ZGK
 
 | | 1.35 angstrom structure of the Kelch domain of Keap1 | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Li, X, Bottoms, C.A, Hannink, M, Beamer, L.J. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conserved solvent and side-chain interactions in the 1.35 Angstrom structure of the Kelch domain of Keap1.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4IL8
 
 | | Crystal structure of an H329A mutant of p. aeruginosa PMM/PGM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Lee, Y, Mehra-Chaudhary, R, Furdui, C, Beamer, L. | | Deposit date: | 2012-12-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an essential active-site residue in the alpha-D-phosphohexomutase enzyme superfamily.
Febs J., 280, 2013
|
|
