3Q8C
 
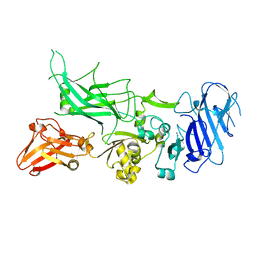 | | Crystal structure of Protective Antigen W346F (pH 5.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8E
 
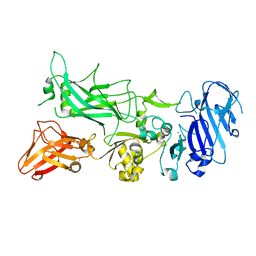 | | Crystal structure of Protective Antigen W346F (pH 8.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8B
 
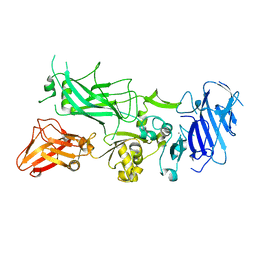 | | Crystal structure of WT Protective Antigen (pH 9.0) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8F
 
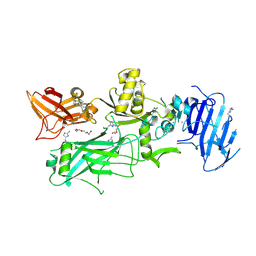 | | Crystal structure of 2-Fluorohistine labeled Protective Antigen (pH 5.8) | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
8TXA
 
 | | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Balsamo, A, Bruno, C, Edele, C, Fabian, T, Jannotta, R, Lee, R, Schryver, D, Warsaw, J, Warsaw, L, Stojanoff, V, Battaile, K, Perez, A, Bolen, R. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum
To Be Published
|
|
3P8K
 
 | | Crystal Structure of a putative carbon-nitrogen family hydrolase from Staphylococcus aureus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gordon, R.D, Qiu, W, Battaile, K, Lam, K, Soloveychik, M, Benetteraj, D, Romanov, V, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-10-14 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of carbon-nitrogen family hydrolase from Staphylococcus aureus
To be Published
|
|
3P8A
 
 | | Crystal Structure of a hypothetical protein from Staphylococcus aureus | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lam, R, Qiu, W, Battaile, K, Lam, K, Romanov, V, Chan, T, Pai, E, Chirgadze, N.Y. | | Deposit date: | 2010-10-13 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a hypothetical protein from Staphylococcus aureus
To be Published
|
|
3R9I
 
 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
8EJX
 
 | |
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
3PZF
 
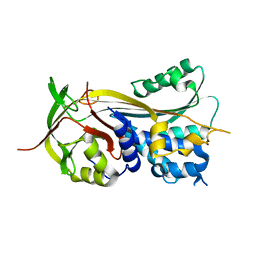 | | 1.75A resolution structure of Serpin-2 from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, An, C, Michel, K. | | Deposit date: | 2010-12-14 | | Release date: | 2011-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of native Anopheles gambiae serpin-2, a negative regulator of melanization in mosquitoes.
Proteins, 79, 2011
|
|
4PEO
 
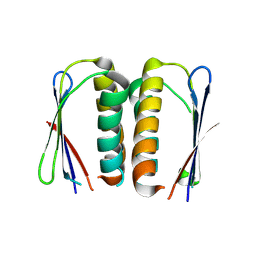 | | Crystal structure of a hypothetical protein from Staphylococcus aureus. | | Descriptor: | Hypothetical protein | | Authors: | McGrath, T.E, Kisselman, G, Romanov, V, Wu-Brown, J, Soloveychik, M, Chan, T.S.Y, Gordon, R.D, Thambipillai, D, Dharamsi, A, Mansoury, K, Battaile, K.P, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of a hypothetical protein from Staphylococcus aureus.
To Be Published
|
|
8G0F
 
 | |
3QH7
 
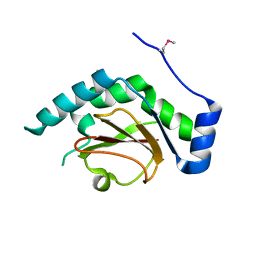 | | 2.5 A resolution structure of Se-Met labeled CT296 from Chlamydia trachomatis | | Descriptor: | CT296 | | Authors: | Kemege, K, Hickey, J, Lovell, S, Battaile, K.P, Zhang, Y, Hefty, P.S. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-05 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes.
J. Bacteriol., 193, 2011
|
|
4O8I
 
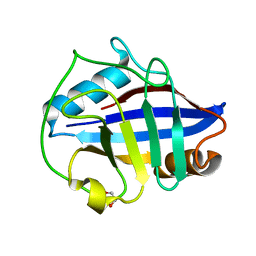 | | 1.45A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
8G0G
 
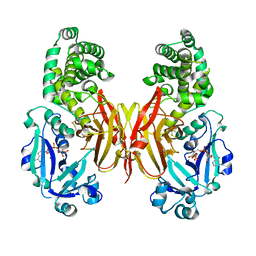 | | Crystal structure of diphtheria toxin H223Q/H257Q double mutant (pH 4.5) | | Descriptor: | ADENYLYL-3'-5'-PHOSPHO-URIDINE-3'-MONOPHOSPHATE, Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Ladokhin, A.S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine Protonation and Conformational Switching in Diphtheria Toxin Translocation Domain.
Toxins, 15, 2023
|
|
4QAS
 
 | | 1.27 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
3R9J
 
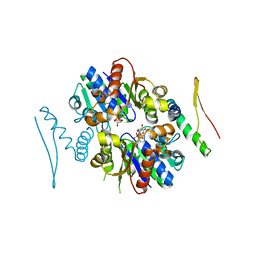 | | 4.3A resolution structure of a MinD-MinE(I24N) protein complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
4QAQ
 
 | | 1.58 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAT
 
 | | 1.75 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis bound to MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QFB
 
 | | 1.99 A resolution structure of SeMet-CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
3QH6
 
 | | 1.8A resolution structure of CT296 from Chlamydia trachomatis | | Descriptor: | CT296, TETRAETHYLENE GLYCOL | | Authors: | Kemege, K, Hickey, J, Lovell, S, Battaile, K.P, Zhang, Y, Hefty, P.S. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes.
J.Bacteriol., 193, 2011
|
|
4QAR
 
 | | 1.45 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis bound to Adenine | | Descriptor: | ADENINE, CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
8TGB
 
 | | Crystal structure of root lateral formation protein (RLF) b5-domain from Oryza sativa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, root lateral formation protein (RLF) | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Benson, D.R. | | Deposit date: | 2023-07-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The N-terminal intrinsically disordered region of Ncb5or docks with the cytochrome b 5 core to form a helical motif that is of ancient origin.
Proteins, 92, 2024
|
|
7TPM
 
 | | Structure of the outer-membrane lipoprotein carrier protein (LolA) from Borrelia burgdorferi | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CHLORIDE ION, Outer membrane lipoprotein carrier protein LolA, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Zueckert, W.R. | | Deposit date: | 2022-01-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the outer-membrane lipoprotein carrier protein (LolA) from Borrelia burgdorferi
To be published
|
|
