6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
6VSJ
 
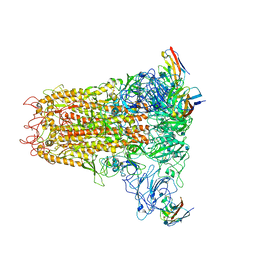 | | Cryo-electron microscopy structure of mouse coronavirus spike protein complexed with its murine receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, Spike glycoprotein | | Authors: | Shang, J, Wan, Y.S, Liu, C, Yount, B, Gully, K, Yang, Y, Auerbach, A, Peng, G.Q, Baric, R, Li, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry.
Plos Pathog., 16, 2020
|
|
4L3N
 
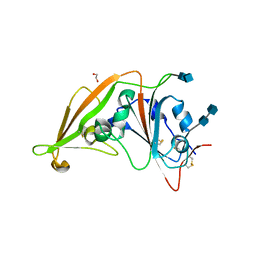 | | Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Chen, Y, Rajashankar, K.R, Yang, Y, Agnihothram, S.S, Liu, C, Lin, Y.-L, Baric, R.S, Li, F. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the receptor-binding domain from newly emerged middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
3EE7
 
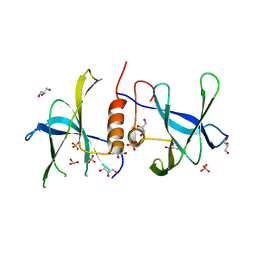 | | Crystal Structure of SARS-CoV nsp9 G104E | | Descriptor: | GLYCEROL, PHOSPHATE ION, Replicase polyprotein 1a | | Authors: | Miknis, Z.J, Donaldson, E.F, Umland, T.C, Rimmer, R, Baric, R.S, Schultz, L.W. | | Deposit date: | 2008-09-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth
J.Virol., 83, 2009
|
|
5H37
 
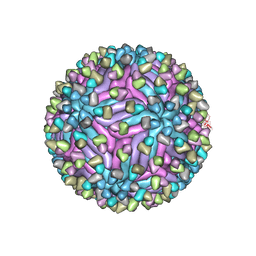 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C10 IgG heavy chain variable region, C10 IgG light chain variable region, ... | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lim, X.-N, Ooi, J.S.G, Lambert, S, Tan, T.Y, Widman, D, Shi, J, Baric, R.S, Lok, S.-M. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
