7RGO
 
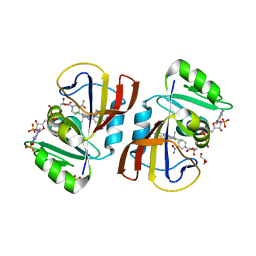 | | DfrA5 complexed with NADPH and 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | 分子名称: | 4'-chloro-3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl][1,1'-biphenyl]-4-carboxamide, Dihydrofolate reductase type 5, GLYCEROL, ... | | 著者 | Lombardo, M.N, Wright, D.L. | | 登録日 | 2021-07-15 | | 公開日 | 2022-05-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7RGJ
 
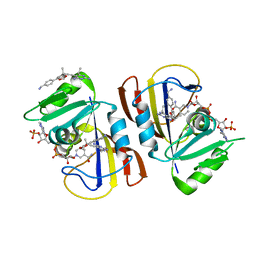 | | DfrA1 complexed with NADPH and 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | 分子名称: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, 5-[(3S)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, CALCIUM ION, ... | | 著者 | Lombardo, M.N, Wright, D.L. | | 登録日 | 2021-07-15 | | 公開日 | 2022-05-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
6UXC
 
 | |
7REG
 
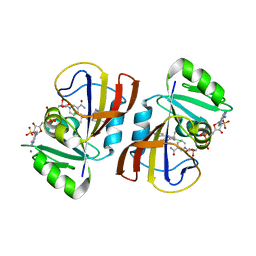 | | DfrA1 complexed with NADPH and 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | 分子名称: | 4'-chloro-3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl][1,1'-biphenyl]-4-carboxamide, CALCIUM ION, Dihydrofolate reductase type 1, ... | | 著者 | Lombardo, M.N, Wright, D.L. | | 登録日 | 2021-07-12 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
6QBT
 
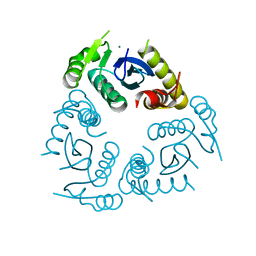 | |
6QBV
 
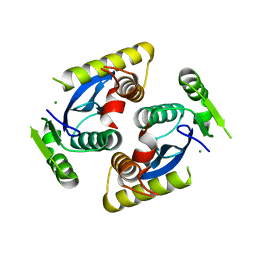 | |
6QBW
 
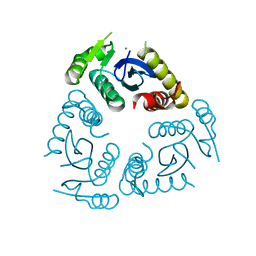 | |
6UXD
 
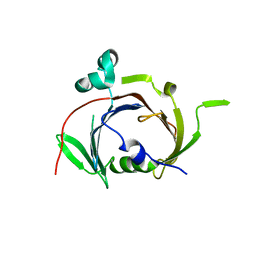 | |
8Q1H
 
 | |
8Q1G
 
 | |
8Q1J
 
 | |
5UE0
 
 | | 1.90 A resolution structure of CT622 C-terminal domain from Chlamydia trachomatis | | 分子名称: | CT622 protein, SULFATE ION | | 著者 | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | 登録日 | 2016-12-29 | | 公開日 | 2018-01-10 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Loss of Expression of a Single Type 3 Effector (CT622) Strongly ReducesChlamydia trachomatisInfectivity and Growth.
Front Cell Infect Microbiol, 8, 2018
|
|
1WA1
 
 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | 著者 | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | 登録日 | 2004-10-22 | | 公開日 | 2005-01-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
1WA0
 
 | | Crystal Structure Of W138H Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | 著者 | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | 登録日 | 2004-10-22 | | 公開日 | 2005-01-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
1WA2
 
 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase with nitrite bound | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE,, ... | | 著者 | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | 登録日 | 2004-10-22 | | 公開日 | 2005-01-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
4MY6
 
 | | EnaH-EVH1 in complex with peptidomimetic low-molecular weight inhibitor Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-OH | | 分子名称: | (3aR,5aS,8S,10aS)-1-[(3S,6R,8aS)-1'-[(2S)-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8a-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-10-oxidanylidene-2,3,3a,5a,8,10a-hexahydrodipyrrolo[3,2-b:3',1'-f]azepine-8-carboxylic acid, BROMIDE ION, Protein enabled homolog | | 著者 | Barone, M, Roske, Y, Kuehne, R. | | 登録日 | 2013-09-27 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A modular toolkit to inhibit proline-rich motif-mediated protein-protein interactions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2CAL
 
 | | Crystal structure of His143Met rusticyanin | | 分子名称: | COPPER (I) ION, RUSTICYANIN | | 著者 | Barrett, M.L, Harvey, I, Sundararajan, M, Surendran, R, Hall, J.F, Ellis, M.J, Hough, M.A, Strange, R.W, Hillier, I.H, Hasnain, S.S. | | 登録日 | 2005-12-21 | | 公開日 | 2006-01-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic Resolution Crystal Structures, Exafs, and Quantum Chemical Studies of Rusticyanin and its Two Mutants Provide Insight Into its Unusual Properties.
Biochemistry, 45, 2006
|
|
2CAK
 
 | | 1.27Angstrom Structure of Rusticyanin from Thiobacillus ferrooxidans | | 分子名称: | COPPER (I) ION, RUSTICYANIN | | 著者 | Barrett, M.L, Harvey, I, Sundararajan, M, Surendran, R, Hall, J.F, Ellis, M.J, Hough, M.A, Strange, R.W, Hillier, I.H, Hasnain, S.S. | | 登録日 | 2005-12-21 | | 公開日 | 2006-03-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Atomic Resolution Crystal Structures, Exafs, and Quantum Chemical Studies of Rusticyanin and its Two Mutants Provide Insight Into its Unusual Properties.
Biochemistry, 45, 2006
|
|
5ECC
 
 | | Klebsiella pneumoniae DfrA1 complexed with NADPH and 6-ethyl-5-(3-(2-methoxy-5-(pyridin-4-yl)phenyl)prop-1-yn-1-yl)pyrimidine-2,4-diamine | | 分子名称: | 6-ethyl-5-{3-[2-methoxy-5-(pyridin-4-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, CALCIUM ION, Dehydrofolate reductase type I, ... | | 著者 | Lombardo, M.N, Anderson, A.C. | | 登録日 | 2015-10-20 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal Structures of Trimethoprim-Resistant DfrA1 Rationalize Potent Inhibition by Propargyl-Linked Antifolates.
ACS Infect Dis, 2, 2016
|
|
2F44
 
 | |
3CV3
 
 | |
2F3N
 
 | |
3CUY
 
 | | Crystal Structure of GumK mutant D157A | | 分子名称: | Glucuronosyltransferase GumK | | 著者 | Barreras, M. | | 登録日 | 2008-04-17 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase.
J.Biol.Chem., 283, 2008
|
|
3TUL
 
 | | Crystal structure of N-terminal region of Type III Secretion Major Translocator SipB (residues 82-226) | | 分子名称: | Cell invasion protein sipB | | 著者 | Barta, M.L, Dickenson, N.E, Patel, M, Keightley, J.A, Picking, W.D, Picking, W.L, Geisbrecht, B.V. | | 登録日 | 2011-09-16 | | 公開日 | 2012-02-15 | | 最終更新日 | 2012-03-28 | | 実験手法 | X-RAY DIFFRACTION (2.793 Å) | | 主引用文献 | The Structures of Coiled-Coil Domains from Type III Secretion System Translocators Reveal Homology to Pore-Forming Toxins.
J.Mol.Biol., 417, 2012
|
|
3R9V
 
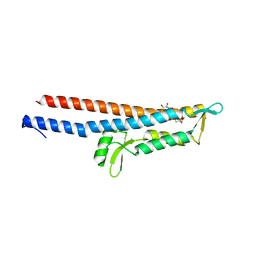 | | Cocrystal Structure of Proteolytically Truncated Form of IpaD from Shigella flexneri Bound to Deoxycholate | | 分子名称: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, Invasin ipaD | | 著者 | Barta, M.L, Dickenson, N.E, Picking, W.L, Picking, W.D, Geisbrecht, B.V. | | 登録日 | 2011-03-26 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of the bile salt binding site on IpaD from Shigella flexneri and the influence of ligand binding on IpaD structure.
Proteins, 80, 2012
|
|
