7U2P
 
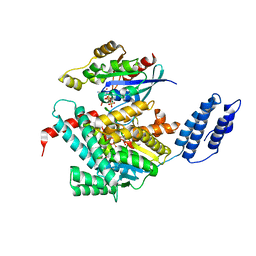 | | Structure of TcdA GTD in complex with RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Glucosyltransferase TcdA, MAGNESIUM ION, ... | | Authors: | Baohua, C, Zheng, L, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of the glucosyltransferase domain of TcdA in complex with RhoA provides insights into substrate recognition.
Sci Rep, 12, 2022
|
|
7U1Z
 
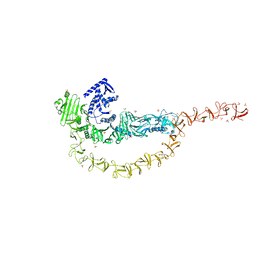 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
6VWN
 
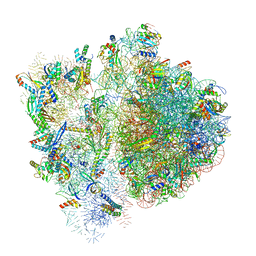 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P-site tRNA (non-rotated conformation, Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
6VWM
 
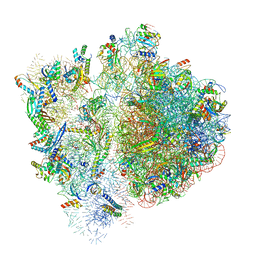 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P-site tRNA (non-rotated conformation, Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
6VWL
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P/E tRNA (rotated conformation) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
8XLD
 
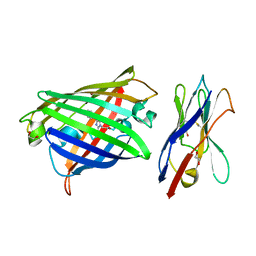 | | Structure of the GFP:GFP-nanobody complex from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Nanobody(Staygold-S2G10)-Nanobody(Staygold-S4F1), ZINC ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the GFP:GFP-nanobody complex from Biortus.
To Be Published
|
|
8XP5
 
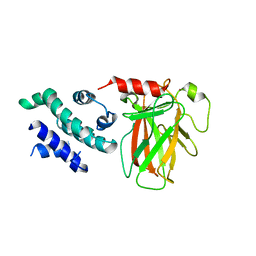 | | The Crystal Structure of p53/BCL-xL fusion complex from Biortus. | | Descriptor: | Bcl-2-like protein 1,Cellular tumor antigen p53, ZINC ION | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2024-01-03 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Crystal Structure of p53/BCL-xL fusion complex from Biortus.
To Be Published
|
|
8XPT
 
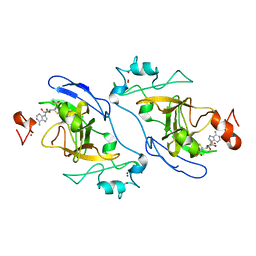 | | The Crystal Structure of EHMT1 from Biortus. | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2024-01-04 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Crystal Structure of EHMT1 from Biortus.
To Be Published
|
|
8XN8
 
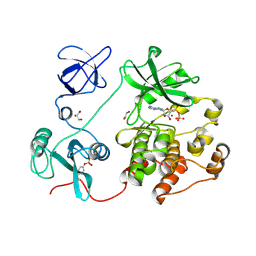 | | The Crystal Structure of SRC from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2023-12-29 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of SRC from Biortus.
To Be Published
|
|
8XI8
 
 | |
8Y9A
 
 | |
8X2Q
 
 | | The Crystal Structure of APC from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Adenomatous polyposis coli protein | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Bao, C. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of APC from Biortus.
To Be Published
|
|
8ZWV
 
 | | The Crystal Structure of carbonic anhydrase II from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 2, ZINC ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Bao, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of carbonic anhydrase II from Biortus.
To Be Published
|
|
8WD3
 
 | | The Crystal Structure of JMJD2A(M1-L359) from Biortus. | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Bao, C. | | Deposit date: | 2023-09-14 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of JMJD2A(M1-L359) from Biortus.
To Be Published
|
|
6AIB
 
 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4R32
 
 | | Crystal Structure Analysis of Pyk2 and Paxillin LD motifs | | Descriptor: | Paxillin, Protein-tyrosine kinase 2-beta | | Authors: | Vanarotti, M, Miller, D.J, Guibao, C.D, Nourse, A, Zheng, J.J. | | Deposit date: | 2014-08-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.505 Å) | | Cite: | Structural and Mechanistic Insights into the Interaction between Pyk2 and Paxillin LD Motifs.
J.Mol.Biol., 426, 2014
|
|
1KTM
 
 | |
2JX0
 
 | | The paxillin-binding domain (PBD) of G Protein Coupled Receptor (GPCR)-kinase (GRK) interacting protein 1 (GIT1) | | Descriptor: | ARF GTPase-activating protein GIT1 | | Authors: | Zhang, Z, Guibao, C.D, Simmerman, J.A, Zheng, J. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs.
J.Biol.Chem., 283, 2008
|
|
6AIC
 
 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2LK4
 
 | |
3U3F
 
 | | Structural basis for the interaction of Pyk2 PAT domain with paxillin LD motifs | | Descriptor: | Paxillin LD2 peptide, Protein-tyrosine kinase 2-beta | | Authors: | Vanarotti, M, Miller, D.J, Guibao, C.C, Zheng, J.J. | | Deposit date: | 2011-10-05 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural and Mechanistic Insights into the Interaction between Pyk2 and Paxillin LD Motifs.
J.Mol.Biol., 426, 2014
|
|
2L6F
 
 | | NMR Solution structure of FAT domain of FAK complexed with LD2 and LD4 motifs of PAXILLIN | | Descriptor: | Focal adhesion kinase 1, linker1, Paxillin, ... | | Authors: | Bertolucci, C.M, Guibao, C, Zhang, C, Zheng, J. | | Deposit date: | 2010-11-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Fat Domain of Fak Complexed with Ld2 and Ld4 Motifs of Paxillin
To be Published
|
|
2L6H
 
 | |
2L6G
 
 | |
