1TJ9
 
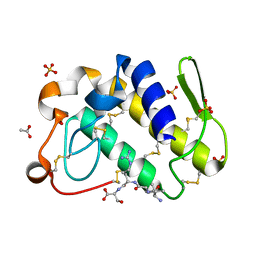 | | Structure of the complexed formed between group II phospholipase A2 and a rationally designed tetra peptide,Val-Ala-Arg-Ser at 1.1A resolution | | Descriptor: | ACETIC ACID, Phospholipase A2, SULFATE ION, ... | | Authors: | Singh, N, Ethayathulla, A.S, K Somvanshi, R, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Kaur, P, Singh, T.P. | | Deposit date: | 2004-06-03 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the complex formed between group II phospholipase A2 and a rationally designed tetra peptide,Val-Ala-Arg-Ser at 1.1A resolution
TO BE PUBLISHED
|
|
2XIC
 
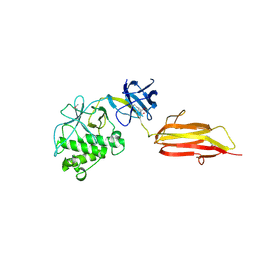 | | Pilus-presented adhesin, Spy0125 (Cpa), P212121 form (ESRF data) | | Descriptor: | ANCILLARY PROTEIN 1 | | Authors: | Pointon, J.A, Smith, W.D, Saalbach, G, Crow, A, Kehoe, M.A, Banfield, M.J. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Highly Unusual Thioester Bond in a Pilus Adhesin is Required for Efficient Host Cell Interaction
J.Biol.Chem., 285, 2010
|
|
1TH6
 
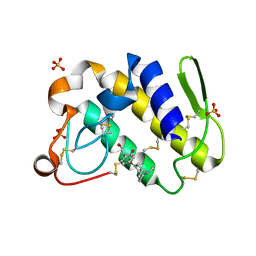 | | Crystal structure of phospholipase A2 in complex with atropine at 1.23A resolution | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, Phospholipase A2, SULFATE ION | | Authors: | Singh, N, Pal, A, Jabeen, T, Sharma, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structure of phospholipase A2 in complex with atropine at 1.23A resolution
To be Published
|
|
1TK4
 
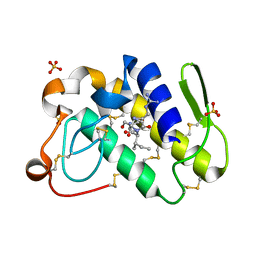 | | Crystal structure of russells viper phospholipase A2 in complex with a specifically designed tetrapeptide Ala-Ile-Arg-Ser at 1.1 A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION, Tetrapeptide Ala-Ile-Arg-Ser | | Authors: | Singh, N, Bilgrami, S, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Kaur, P, Singh, T.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of russells viper phospholipase A2 with a specifically designed tetrapeptide Ala-Ile-Arg-Ser at 1.1 A resolution
TO BE PUBLISHED
|
|
2XV3
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAAAHAAAAM), chemically reduced, pH5.3 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
2XID
 
 | | Pilus-presented adhesin, Spy0125 (Cpa), P212121 form (DLS) | | Descriptor: | ANCILLARY PROTEIN 1 | | Authors: | Pointon, J.A, Smith, W.D, Saalbach, G, Crow, A, Kehoe, M.A, Banfield, M.J. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Highly Unusual Thioester Bond in a Pilus Adhesin Required for Efficient Host Cell Interaction
J.Biol.Chem., 285, 2010
|
|
2Q1P
 
 | | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution | | Descriptor: | N-PROPANOL, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Kumar, S, Hariprasad, G, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-05-25 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution
To be Published
|
|
1WKO
 
 | |
3ZVD
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 83 | | Descriptor: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
1TD7
 
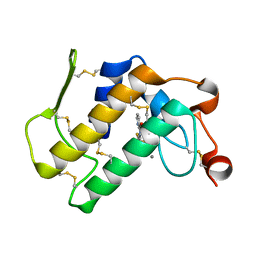 | | Interactions of a specific non-steroidal anti-inflammatory drug (NSAID) with group I phospholipase A2 (PLA2): Crystal structure of the complex formed between PLA2 and niflumic acid at 2.5 A resolution | | Descriptor: | 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Jabeen, T, Singh, N, Singh, R.K, Sharma, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-steroidal anti-inflammatory drugs as potent inhibitors of phospholipase A2: structure of the complex of phospholipase A2 with niflumic acid at 2.5 Angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1TJK
 
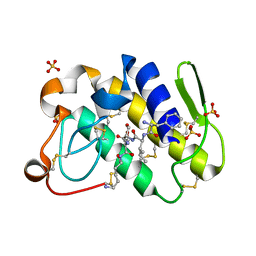 | | Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe- Leu- Ser- Thr- Lys at 1.2 A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Jabeen, T, Somvanshi, R.K, Sharma, S, Perbandt, M, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2004-06-06 | | Release date: | 2004-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe - Leu - Ser - Thr - Lys at 1.2 A resolution
To be Published
|
|
1TDV
 
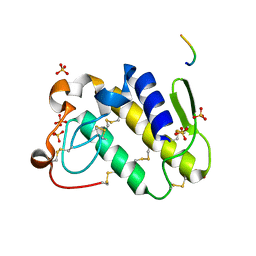 | | Non-specific binding to phospholipase A2:Crystal structure of the complex of PLA2 with a designed peptide Tyr-Trp-Ala-Ala-Ala-Ala at 1.7A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION, YWAAAA | | Authors: | Singh, N, Jabeen, T, Ethayathulla, A.S, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-24 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-specific binding to phospholipase A2:Crystal structure of the complex of PLA2 with a designed peptide Tyr-Trp-Ala-Ala-Ala-Ala at 1.7A resolution
to be published
|
|
1SKU
 
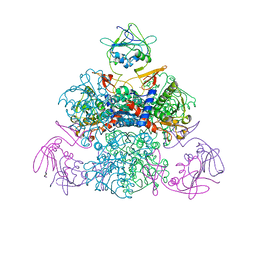 | | E. coli Aspartate Transcarbamylase 240's Loop Mutant (K244N) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, MALONATE ION, ... | | Authors: | Alam, N, Stieglitz, K.A, Caban, M.D, Gourinath, S, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 240s Loop Interactions Stabilize the T State of Escherichia coli Aspartate Transcarbamoylase.
J.Biol.Chem., 279, 2004
|
|
2L4T
 
 | | GIP/Glutaminase L peptide complex | | Descriptor: | Glutaminase L peptide, Tax1-binding protein 3 | | Authors: | Zoetewey, D.L, Ovee, M, Banerjee, M, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous binding at the crossroads of numerous cancer pathways: insight from the binding of glutaminase interacting protein with glutaminase L.
Biochemistry, 50, 2011
|
|
1SKG
 
 | | Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser | | Descriptor: | METHANOL, Phospholipase A2, SULFATE ION, ... | | Authors: | Ethayathulla, A.S, Singh, N, Sharma, S, Makker, J, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser
To be Published
|
|
4F8C
 
 | |
2O1L
 
 | | Structure of a complex of C-terminal lobe of bovine lactoferrin with disaccharide at 1.97 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, N, Sharma, S, Perbandt, M, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of a complex of C-terminal lobe of bovine lactoferrin with disaccharide at 1.97 A resolution
To be Published
|
|
1WKP
 
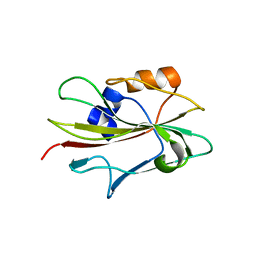 | | Flowering locus t (ft) from arabidopsis thaliana | | Descriptor: | FLOWERING LOCUS T protein, SULFATE ION | | Authors: | Miller, D, Banfield, M.J, Winter, V.J, Brady, R.L. | | Deposit date: | 2004-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A divergent external loop confers antagonistic activity on floral regulators FT and TFL1.
Embo J., 25, 2006
|
|
2L4S
 
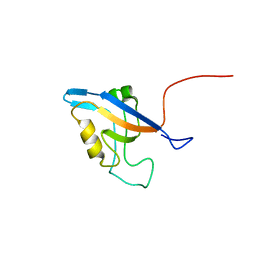 | | Promiscuous Binding at the Crossroads of Numerous Cancer Pathways: Insight from the Binding of GIP with Glutaminase L | | Descriptor: | Tax1-binding protein 3 | | Authors: | Zoetewey, D.L, Ovee, M, Banerjee, M, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous binding at the crossroads of numerous cancer pathways: insight from the binding of glutaminase interacting protein with glutaminase L.
Biochemistry, 50, 2011
|
|
1KN3
 
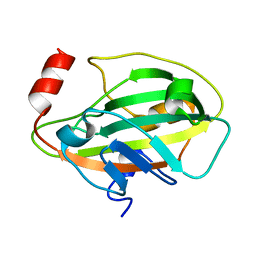 | |
1IC6
 
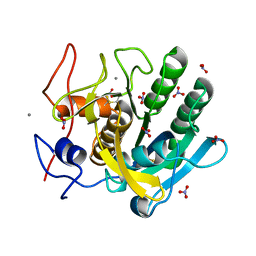 | | STRUCTURE OF A SERINE PROTEASE PROTEINASE K FROM TRITIRACHIUM ALBUM LIMBER AT 0.98 A RESOLUTION | | Descriptor: | CALCIUM ION, NITRATE ION, PROTEINASE K | | Authors: | Betzel, C, Gourinath, S, Kumar, P, Kaur, P, Perbandt, M, Eschenburg, S, Singh, T.P. | | Deposit date: | 2001-03-30 | | Release date: | 2001-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 A resolution.
Biochemistry, 40, 2001
|
|
1CE7
 
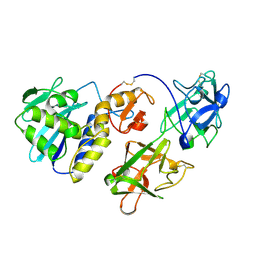 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
2W89
 
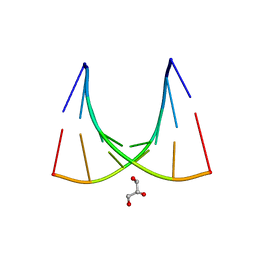 | | Crystal structure of the E.coli tRNAArg aminoacyl stem issoacceptor RR-1660 at 2.0 Angstroem resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*CP*CP*GP)-3', GLYCEROL | | Authors: | Eichert, A, Schreiber, A, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E. Coli tRNA(Arg) Aminoacyl Stem Isoacceptor Rr-1660 at 2.0 A Resolution.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
4B6X
 
 | | Crystal structure of in planta processed AvrRps4 | | Descriptor: | ACETATE ION, AVIRULENCE PROTEIN | | Authors: | Sohn, K.H, Hughes, R.K, Piquerez, S.J, Jones, J.D.G, Banfield, M.J. | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct Regions of the Pseudomonas Syringae Coiled-Coil Effector Avrrps4 are Required for Activation of Immunity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2G58
 
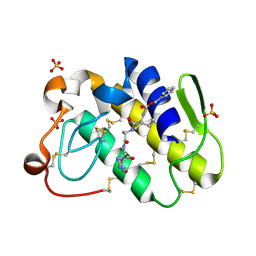 | | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution | | Descriptor: | (PHQ)IARS, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Prem Kumar, R, Singh, N, Somvanshi, R.K, Ethayathulla, A.S, Dey, S, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution
To be Published
|
|
