1Y6U
 
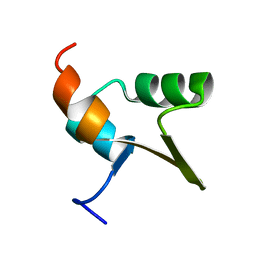 | |
1J5H
 
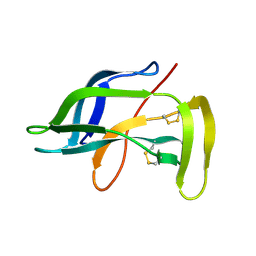 | | Solution Structure of Apo-Neocarzinostatin | | Descriptor: | Apo-Neocarzinostatin | | Authors: | Urbaniak, M.D, Muskett, F.W, Finucane, M.D, Caddick, S, Woolfson, D.N. | | Deposit date: | 2002-05-02 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore
Biochemistry, 41, 2002
|
|
1H5Y
 
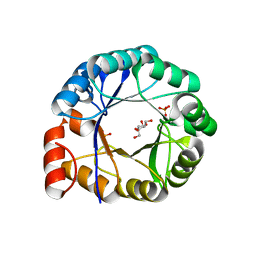 | | HisF protein from Pyrobaculum aerophilum | | Descriptor: | GLYCEROL, HISF, PHOSPHATE ION | | Authors: | Banfield, M.J, Lott, J.S, McCarthy, A.A, Baker, E.N. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hisf, a Histidine Biosynthetic Protein from Pyrobaculum Aerophilum
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2FTA
 
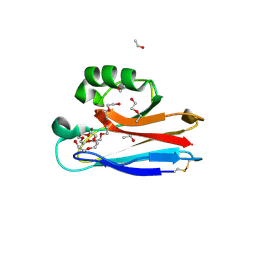 | |
5JPC
 
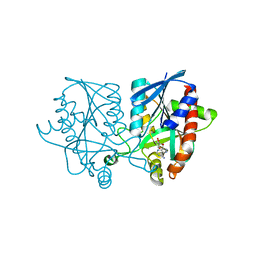 | | Joint X-ray/neutron structure of MTAN complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Aminodeoxyfutalosine nucleosidase | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
439D
 
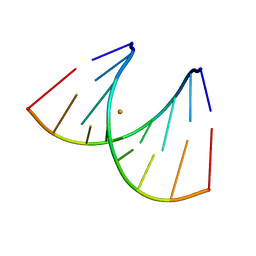 | | 5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3', 5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3' | | Descriptor: | BARIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3') | | Authors: | Perbandt, M, Lorenz, S, Vallazza, M, Erdmann, V.A, Betzel, C. | | Deposit date: | 1999-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1J5I
 
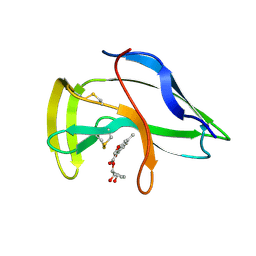 | | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore | | Descriptor: | 2-HYDROXY-7-METHOXY-5-METHYL-NAPHTHALENE-1-CARBOXYLIC ACID MESO-2,5-DIHYDROXY-CYCLOPENT-3-ENYL ESTER, PROTEIN (Apo-Neocarzinostatin) | | Authors: | Urbaniak, M.D, Muskett, F.W, Finucane, M.D, Caddick, S, Woolfson, D.N. | | Deposit date: | 2002-05-02 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore
Biochemistry, 41, 2002
|
|
6ITB
 
 | |
1BD9
 
 | |
1BEH
 
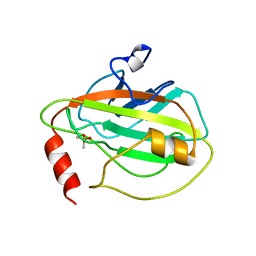 | | HUMAN PHOSPHATIDYLETHANOLAMINE BINDING PROTEIN IN COMPLEX WITH CACODYLATE | | Descriptor: | CACODYLATE ION, PHOSPHATIDYLETHANOLAMINE BINDING PROTEIN | | Authors: | Banfield, M.J, Barker, J.J, Perry, A, Brady, R.L. | | Deposit date: | 1998-05-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction.
Structure, 6, 1998
|
|
1AOK
 
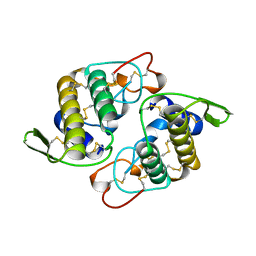 | | VIPOXIN COMPLEX | | Descriptor: | ACETATE ION, VIPOXIN COMPLEX | | Authors: | Perbandt, M, Wilson, J.C, Eschenburg, S, Betzel, C. | | Deposit date: | 1997-07-07 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of vipoxin at 2.0 A: an example of regulation of a toxic function generated by molecular evolution.
FEBS Lett., 412, 1997
|
|
6W18
 
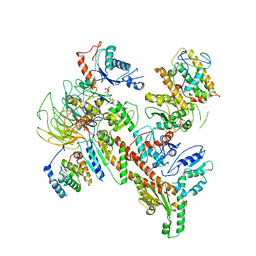 | | Structure of S. pombe Arp2/3 complex in inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1, ... | | Authors: | Shaaban, M, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1CLO
 
 | |
6ITF
 
 | |
1AE6
 
 | | IGG-FAB FRAGMENT OF MOUSE MONOCLONAL ANTIBODY CTM01 | | Descriptor: | IGG CTM01 FAB (HEAVY CHAIN), IGG CTM01 FAB (LIGHT CHAIN) | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-03-06 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
1AD0
 
 | | FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY A5B7 | | Descriptor: | ANTIBODY A5B7 (HEAVY CHAIN), ANTIBODY A5B7 (LIGHT CHAIN) | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-02-19 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
2FT6
 
 | |
2FT8
 
 | |
1QOU
 
 | |
1OQS
 
 | | Crystal Structure of RV4/RV7 Complex | | Descriptor: | Phospholipase A2 RV-4, Phospholipase A2 RV-7 | | Authors: | Perbandt, M, Betzel, C. | | Deposit date: | 2003-03-11 | | Release date: | 2003-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the heterodimeric neurotoxic complex viperotoxin F (RV-4/RV-7) from the venom of Vipera russelli formosensis at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2FT7
 
 | |
1LNL
 
 | | Structure of deoxygenated hemocyanin from Rapana thomasiana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, hemocyanin | | Authors: | Perbandt, M, Guthoehrlein, E.W, Rypniewski, W, Idakieva, K, Stoeva, S, Voelter, W, Genov, N, Betzel, C. | | Deposit date: | 2002-05-03 | | Release date: | 2003-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity
Biochemistry, 42, 2003
|
|
2F6C
 
 | | Reaction geometry and thermostability of pyranose 2-oxidase from the white-rot fungus Peniophora sp., Thermostability mutant E542K | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase, ... | | Authors: | Bannwarth, M, Heckmann-Pohl, D.M, Bastian, S, Giffhorn, F, Schulz, G.E. | | Deposit date: | 2005-11-29 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Reaction Geometry and Thermostable Variant of Pyranose 2-Oxidase from the White-Rot Fungus Peniophora sp.
Biochemistry, 45, 2006
|
|
5KB3
 
 | | 1.4 A resolution structure of Helicobacter Pylori MTAN in complexed with p-ClPh-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, MAGNESIUM ION | | Authors: | Banco, M.T, Ronning, D.R. | | Deposit date: | 2016-06-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K1Z
 
 | | Joint X-ray/neutron structure of MTAN complex with p-ClPh-Thio-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2016-05-18 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (2.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
