6R1I
 
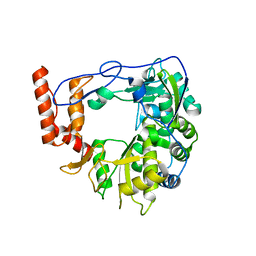 | | Structure of porcine Aichi virus polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
6QWT
 
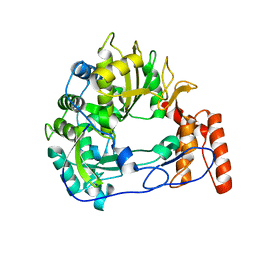 | | Sicinivirus 3Dpol RNA dependent RNA polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
6N0V
 
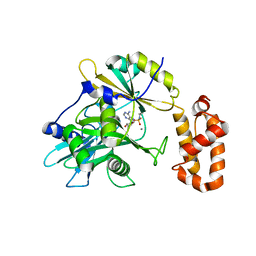 | | tRNA ligase | | Descriptor: | MANGANESE (II) ION, tRNA ligase | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
7OZ3
 
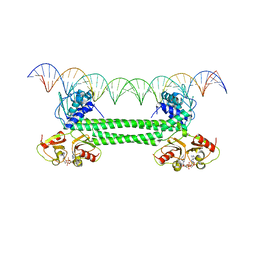 | | S. agalactiae BusR in complex with its busA-promotor DNA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GntR family transcriptional regulator, pBusA_for, ... | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
6ZCF
 
 | |
6ZCG
 
 | |
6ZCH
 
 | |
7QVB
 
 | |
6N0T
 
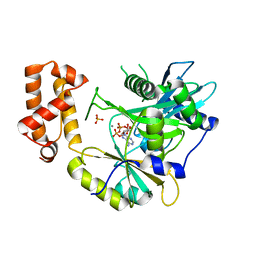 | | tRNA ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
7B5W
 
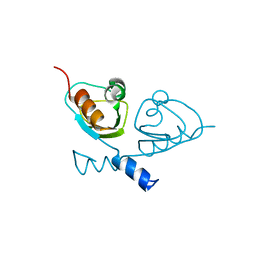 | |
7B5U
 
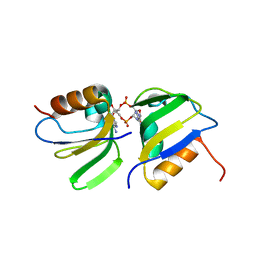 | | RCK_C domain dimer of S.agalactiae BusR in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GntR family transcriptional regulator | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
7B5Y
 
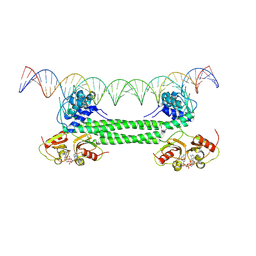 | | S. agalactiae BusR in complex with its busAB-promotor DNA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, BusR binding site in the busAB promotor. strand1, BusR binding site in the busAB promotor. strand2, ... | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
7B5T
 
 | | S. agalactiae BusR transcription factor | | Descriptor: | GntR family transcriptional regulator | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-08-11 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
8H2N
 
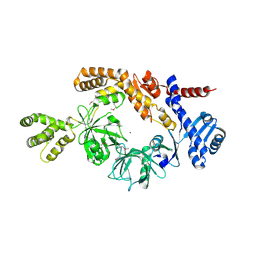 | |
6FX3
 
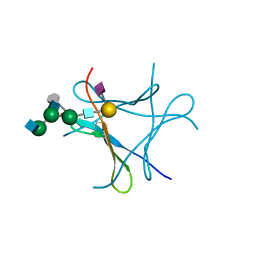 | | crystal structure of Pholiota squarrosa lectin in complex with a dodecasaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, lectin | | Authors: | Cabanettes, A, Varrot, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of Complex Core-Fucosylated N-Glycans by a Mini Lectin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6FX1
 
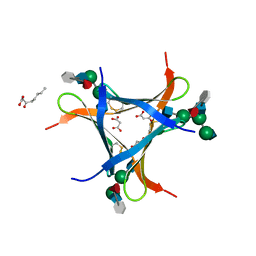 | | Crystal structure of Pholiota squarrosa lectin in complex with an octasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, ... | | Authors: | Cabanettes, A, Varrot, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of Complex Core-Fucosylated N-Glycans by a Mini Lectin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6FX2
 
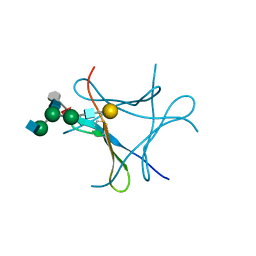 | | crystal structure of Pholiota squarrosa lectin in complex with a decasaccharide | | Descriptor: | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, lectin | | Authors: | Cabanettes, A, Varrot, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of Complex Core-Fucosylated N-Glycans by a Mini Lectin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8H2M
 
 | |
6EKE
 
 | |
5FHL
 
 | |
3GPU
 
 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC4-loop deletion complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*AP*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Banerjee, A, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
3GQ3
 
 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC5-loop deletion complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Banerjee, A, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
7V0N
 
 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21 | | Descriptor: | IgG 21 Fab heavy chain, IgG 21 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7V0O
 
 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94 | | Descriptor: | IgG 94 Fab heavy chain, IgG 94 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7V0P
 
 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a potently neutralizing human antibody IgG-106 | | Descriptor: | IgG 106 Fab heavy chain, IgG 106 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
