5TKV
 
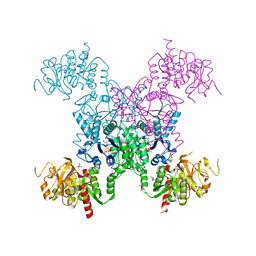 | | X-RAY CRYSTAL STRUCTURE OF THE "CLOSED" CONFORMATION OF CTP-INHIBITED E. COLI CYTIDINE TRIPHOSPHATE (CTP) SYNTHETASE | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, ... | | 著者 | Baldwin, E.P, Endrizzi, J.A. | | 登録日 | 2016-10-07 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1PVP
 
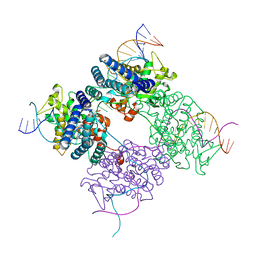 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, ALSHG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | 分子名称: | 34-MER, Recombinase cre | | 著者 | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | 登録日 | 2003-06-28 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVQ
 
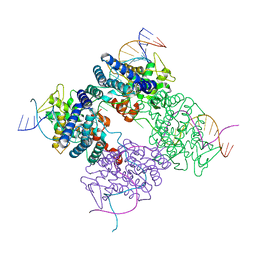 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | 分子名称: | DNA 34-MER, Recombinase cre | | 著者 | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | 登録日 | 2003-06-28 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVR
 
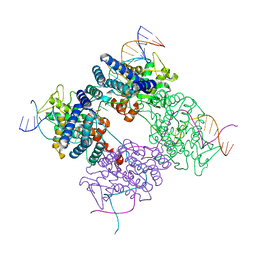 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE LOXP (WILDTYPE) RECOGNITION SITE | | 分子名称: | 34-MER, Recombinase CRE | | 著者 | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | 登録日 | 2003-06-28 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
227L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
222L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
229L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GUANIDINE, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-26 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
252L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-10-28 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1F44
 
 | | CRYSTAL STRUCTURE OF TRIMERIC CRE RECOMBINASE-LOX COMPLEX | | 分子名称: | CRE RECOMBINASE, DNA (5'- D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3') | | 著者 | Baldwin, E.P, Woods, K.C. | | 登録日 | 2000-06-07 | | 公開日 | 2001-10-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction.
J.Mol.Biol., 313, 2001
|
|
220L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
225L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
228L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
223L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
2AD5
 
 | | Mechanisms of feedback regulation and drug resistance of CTP synthetases: structure of the E. coli CTPS/CTP complex at 2.8-Angstrom resolution. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, ... | | 著者 | Endrizzi, J.A, Kim, H, Anderson, P.M, Baldwin, E.P. | | 登録日 | 2005-07-19 | | 公開日 | 2005-11-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanisms of Product Feedback Regulation and Drug Resistance in Cytidine Triphosphate Synthetases from the Structure of a CTP-Inhibited Complex(,).
Biochemistry, 44, 2005
|
|
1S1M
 
 | | Crystal Structure of E. Coli CTP Synthetase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, IODIDE ION, ... | | 著者 | Endrizzi, J.A, Kim, H, Anderson, P.M, Baldwin, E.P. | | 登録日 | 2004-01-06 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Escherichia coli Cytidine Triphosphate Synthetase, a Nucleotide-Regulated Glutamine Amidotransferase/ATP-Dependent Amidoligase Fusion Protein and Homologue of Anticancer and Antiparasitic Drug Targets
Biochemistry, 43, 2004
|
|
1KBU
 
 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | 分子名称: | CRE RECOMBINASE, LOXP | | 著者 | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | 登録日 | 2001-11-06 | | 公開日 | 2002-06-07 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
1MA7
 
 | |
1I6S
 
 | |
1DRG
 
 | | CRYSTAL STRUCTURE OF TRIMERIC CRE RECOMBINASE-LOX COMPLEX | | 分子名称: | 5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3', 5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*GP*C)-3', CRE RECOMBINASE | | 著者 | Woods, K.C, Baldwin, E.P. | | 登録日 | 2000-01-06 | | 公開日 | 2001-10-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction
J.Mol.Biol., 313, 2001
|
|
