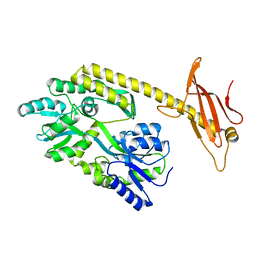
6K7D
 
 | |
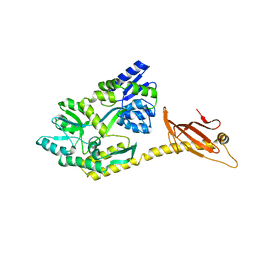
6K7E
 
 | |
6K7F
 
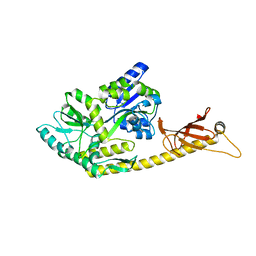 | | Crystal structure of MBPholo-Tim21 fusion protein with a 17-residue helical linker | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Mitochondrial import inner membrane translocase subunit TIM21, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bala, S, Shimada, A, Kohda, D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6K8Q
 
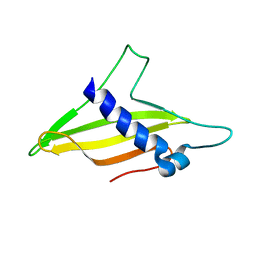 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
8U5Y
 
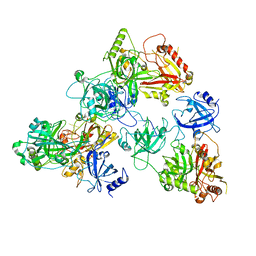 | | human RADX trimer bound to ssDNA | | Descriptor: | DNA (25-MER), RPA-related protein RADX | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Biorxiv, 2023
|
|
8U61
 
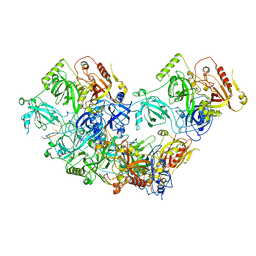 | | Human RADX tetramer bound to ssDNA | | Descriptor: | RPA-related protein RADX, dT25 DNA (25-MER) | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2K9C
 
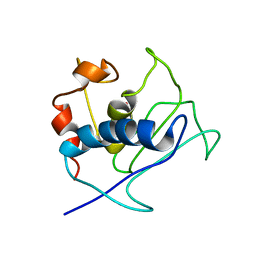 | | Paramagnetic shifts in solid-state NMR of Proteins to elicit structural information | | Descriptor: | COBALT (II) ION, Macrophage metalloelastase | | Authors: | Balayssac, S, Bertini, I, Bhaumik, A, Lelli, M, Luchinat, C. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic shifts in solid-state NMR of proteins to elicit structural information
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5XE4
 
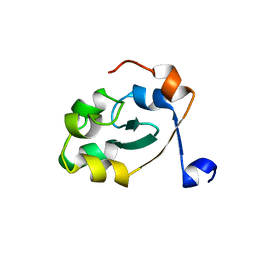 | |
5XEE
 
 | |
8VBZ
 
 | |
8VEP
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 acylated by piperacillin | | Descriptor: | DI(HYDROXYETHYL)ETHER, Piperacillin (Open Form), Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
8VEQ
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 in complex with azlocillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-{[(2R)-2-{[(2-oxoimidazolidin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}ethyl]-1,3-thiazolidine-4-carboxylic acid, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
8VEN
 
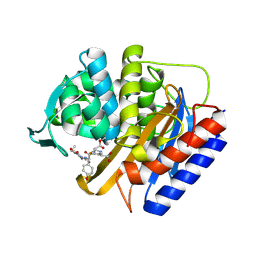 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 in complex with cefoperazone | | Descriptor: | (2R,4R)-2-[(1R)-1-{[(2R)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]-2-(4-hydroxyphenyl)acetyl]amino}-2-oxoethyl]-5-methylidene-1,3-thiazinane-4-carboxylic acid, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
4XK0
 
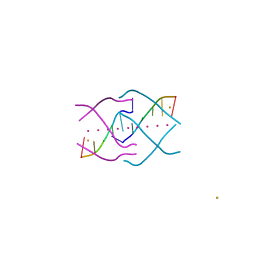 | | Crystal structure of a tetramolecular RNA G-quadruplex in potassium | | Descriptor: | BARIUM ION, POTASSIUM ION, RNA (5'-(*UP*GP*GP*GP*GP*U)-3') | | Authors: | Chen, M.C, Murat, P, Abecassis, K.A, Ferre-D'Amare, A.R, Balasubramanian, S. | | Deposit date: | 2015-01-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase.
Nucleic Acids Res., 43, 2015
|
|
3ZOB
 
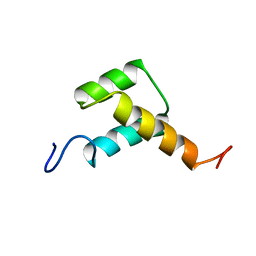 | | Solution structure of chicken Engrailed 2 homeodomain | | Descriptor: | HOMEOBOX PROTEIN ENGRAILED-2 | | Authors: | Carlier, L, Balayssac, S, Cantrelle, F.X, Khemtemourian, L, Chassaing, G, Joliot, A, Lequin, O. | | Deposit date: | 2013-02-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Investigation of Homeodomain Membrane Translocation Properties: Insights from the Structure Determination of Engrailed-2 Homeodomain in Aqueous and Membrane-Mimetic Environments.
Biophys.J., 105, 2013
|
|
1C7W
 
 | | NMR SOLUTION STRUCTURE OF THE CALCIUM-BOUND C-TERMINAL DOMAIN (W81-S161) OF CALCIUM VECTOR PROTEIN FROM AMPHIOXUS | | Descriptor: | CALCIUM VECTOR PROTEIN | | Authors: | Theret, I, Baladi, S, Cox, J.A, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2000-03-27 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequential calcium binding to the regulatory domain of calcium vector protein reveals functional asymmetry and a novel mode of structural rearrangement.
Biochemistry, 39, 2000
|
|
1C7V
 
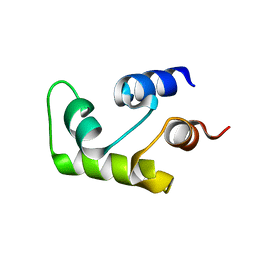 | | NMR SOLUTION STRUCTURE OF THE CALCIUM-BOUND C-TERMINAL DOMAIN (W81-S161) OF CALCIUM VECTOR PROTEIN FROM AMPHIOXUS | | Descriptor: | CALCIUM VECTOR PROTEIN | | Authors: | Theret, I, Baladi, S, Cox, J.A, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2000-03-27 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequential calcium binding to the regulatory domain of calcium vector protein reveals functional asymmetry and a novel mode of structural rearrangement.
Biochemistry, 39, 2000
|
|
4QKK
 
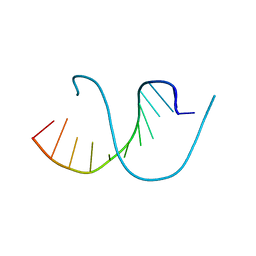 | | Crystal structure of an oligonucleotide containing 5-formylcytosine | | Descriptor: | DNA (5'-D(*CP*TP*AP*(5FC)P*GP*(5FC)P*GP*(5FC)P*GP*TP*AP*G)-3') | | Authors: | Raiber, E.-A, Murat, P, Chirgadze, D.Y, Luisi, B.F, Balasubramanian, S. | | Deposit date: | 2014-06-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 5-Formylcytosine alters the structure of the DNA double helix.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4BWB
 
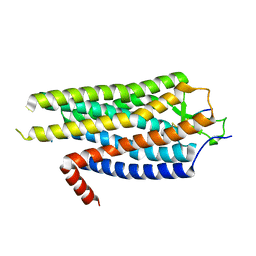 | | Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN, NEUROTENSIN RECEPTOR TYPE 1 | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-07-01 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1J7R
 
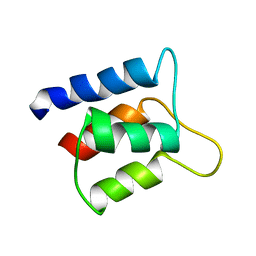 | | Solution structure and backbone dynamics of the defunct EF-hand domain of Calcium Vector Protein | | Descriptor: | Calcium Vector Protein | | Authors: | Theret, I, Baladi, S, Cox, J.A, Gallay, J, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2001-05-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the defunct domain of calcium vector protein.
Biochemistry, 40, 2001
|
|
4BV0
 
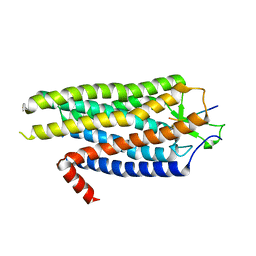 | | High Resolution Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Schuetz, M, Plueckthun, A. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1J7Q
 
 | | Solution structure and backbone dynamics of the defunct EF-hand domain of Calcium Vector Protein | | Descriptor: | Calcium Vector Protein | | Authors: | Theret, I, Baladi, S, Cox, J.A, Gallay, J, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2001-05-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the defunct domain of calcium vector protein.
Biochemistry, 40, 2001
|
|
8S9W
 
 | | Murine S100A7/S100A15 in presence of calcium | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Harrison, S.A, Naretto, A, Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-03-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Comparative analysis of the physical properties of murine and human S100A7: Insight into why zinc piracy is mediated by human but not murine S100A7.
J.Biol.Chem., 299, 2023
|
|
2IVW
 
 | | The solution structure of a domain from the Neisseria meningitidis PilP pilot protein. | | Descriptor: | PILP PILOT PROTEIN | | Authors: | Golovanov, A.P, Balasingham, S, Tzitzilonis, C, Goult, B.T, Lian, L.-Y, Homberset, H, Tonjum, T, Derrick, J.P. | | Deposit date: | 2006-06-20 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a domain from the Neisseria meningitidis lipoprotein PilP reveals a new beta-sandwich fold.
J. Mol. Biol., 364, 2006
|
|
6AC7
 
 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
