4M76
 
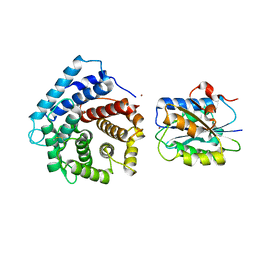 | | Integrin I domain of complement receptor 3 in complex with C3d | | Descriptor: | Complement C3, Integrin alpha-M, NICKEL (II) ION | | Authors: | Bajic, G, Yatime, L, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8VIA
 
 | |
8EQF
 
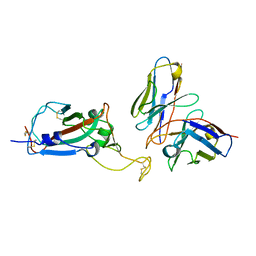 | | cryoEM structure of a broadly neutralizing anti-SARS-CoV-2 antibody STI-9167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Bajic, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | An in vitro experimental pipeline to characterize the epitope of a SARS-CoV-2 neutralizing antibody.
Mbio, 15, 2024
|
|
6UR5
 
 | |
4XW2
 
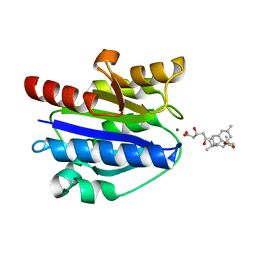 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | Descriptor: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | Authors: | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2015-01-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
6WHK
 
 | | Minimally mutated anti-influenza broadly neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Fab Heavy Chain, ... | | Authors: | Bajic, G, Schmidt, A.G. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Minimally mutated anti-influenza broadly neutralizing antibody
To Be Published
|
|
4HW5
 
 | |
4HWJ
 
 | |
6N5E
 
 | |
6N5D
 
 | |
6N5B
 
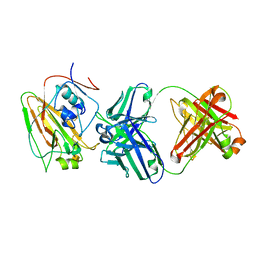 | | Broadly protective antibodies directed to a subdominant influenza hemagglutinin epitope | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Hemagglutinin, antibody heavy chain, ... | | Authors: | Bajic, G, Maron, M.J, Schmidt, A.G. | | Deposit date: | 2018-11-21 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope.
Cell Host Microbe, 25, 2019
|
|
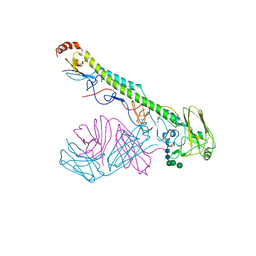
7JIX
 
 | |
7KQH
 
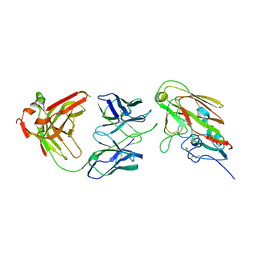 | |
7KQG
 
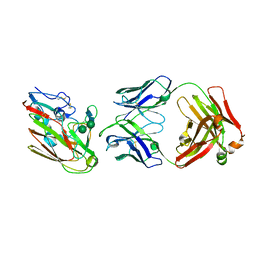 | |
7KQI
 
 | |
7RBV
 
 | | SARS-CoV-2 Spike in complex with PVI.V6-14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PVI.V6-14 Fab heavy chain, ... | | Authors: | Altomare, C.G, Bajic, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a Vaccine-Induced, Germline-Encoded Human Antibody Defines a Neutralizing Epitope on the SARS-CoV-2 Spike N-Terminal Domain.
Mbio, 13, 2022
|
|
7RBU
 
 | | SARS-CoV-2 Spike in complex with PVI.V6-14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PVI.V6-14 Fab heavy chain, ... | | Authors: | Altomare, C.G, Bajic, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a Vaccine-Induced, Germline-Encoded Human Antibody Defines a Neutralizing Epitope on the SARS-CoV-2 Spike N-Terminal Domain.
Mbio, 13, 2022
|
|
7N62
 
 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | Authors: | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
