4TYP
 
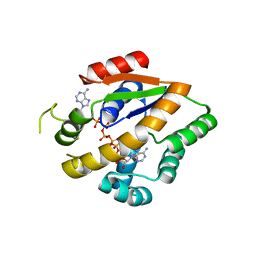 | | Crystal structure of an adenylate kinase mutant--AKm1 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-07-09 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of thermally stable adenylate kinase mutants designed by local structural entropy optimization and structure-guided mutagenesis
J KOREAN SOC APPL BIOL CHEM, 57, 2014
|
|
4TYQ
 
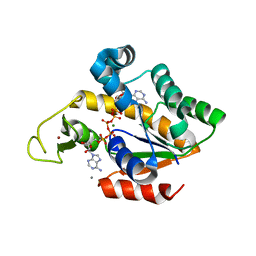 | | Crystal structure of an adenylate kinase mutant--AKm2 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CALCIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-07-09 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of thermally stable adenylate kinase mutants designed by local structural entropy optimization and structure-guided mutagenesis
J KOREAN SOC APPL BIOL CHEM, 57, 2014
|
|
8HEK
 
 | |
8HEL
 
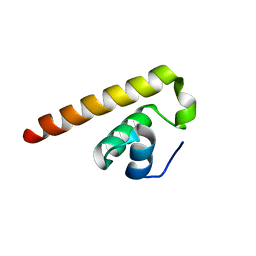 | |
7VZM
 
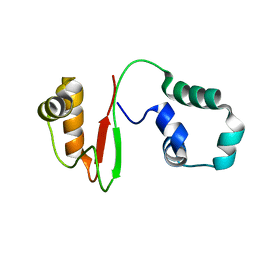 | | Anti-CRISPR AcrIE4-F7 | | Descriptor: | AcrIE4-F7 | | Authors: | Hong, S.H, Lee, G, Bae, E, Suh, J.Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of AcrIE4-F7 reveals a common strategy for dual CRISPR inhibition by targeting PAM recognition sites.
Nucleic Acids Res., 50, 2022
|
|
6PJW
 
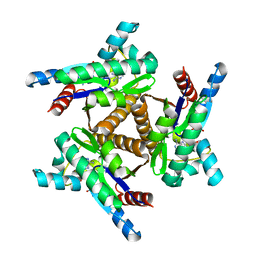 | | Adenylate kinase from Methanococcus igneus - AMP bound form | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylate kinase, MAGNESIUM ION | | Authors: | Moon, S, Kim, J, Olmos, J.L, Bae, E, Phillips Jr, G.N. | | Deposit date: | 2019-06-28 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adenylate kinase from Methanococcus igneus - AMP bound form
To Be Published
|
|
6PK5
 
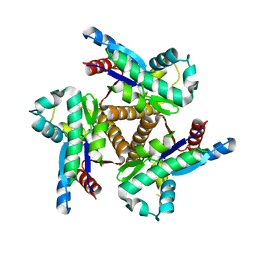 | |
6PSP
 
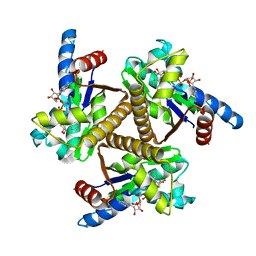 | |
5H1P
 
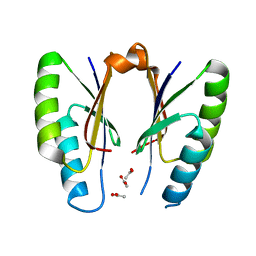 | | CRISPR-associated protein | | Descriptor: | ACETATE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Ka, D, Jeong, U, Bae, E. | | Deposit date: | 2016-10-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity
Struct Dyn, 4, 2017
|
|
5H1O
 
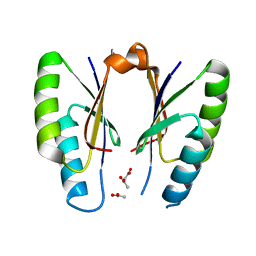 | | CRISPR-associated protein | | Descriptor: | ACETATE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Ka, D, Jeong, U, Bae, E. | | Deposit date: | 2016-10-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity
Struct Dyn, 4, 2017
|
|
2EXR
 
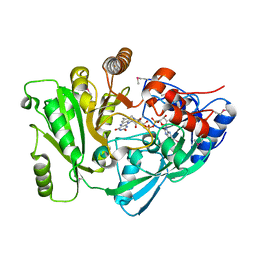 | | X-Ray Structure of Cytokinin Oxidase/Dehydrogenase (CKX) From Arabidopsis Thaliana AT5G21482 | | Descriptor: | Cytokinin dehydrogenase 7, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-11-08 | | Release date: | 2005-11-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase.
Proteins, 70, 2008
|
|
6LKF
 
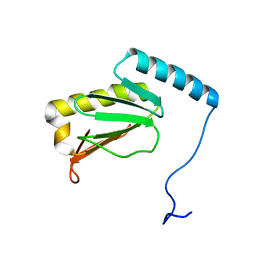 | |
6M3N
 
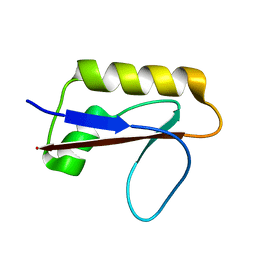 | | Solution structure of anti-CRISPR AcrIF7 | | Descriptor: | anti-CRIPSR AcrIF7 | | Authors: | Kim, I, An, S.Y, Koo, J, Bae, E, Suh, J.Y. | | Deposit date: | 2020-03-04 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into the CRISPR inhibition of AcrIF7.
Nucleic Acids Res., 48, 2020
|
|
4ZKJ
 
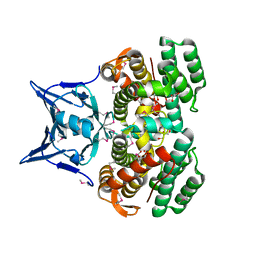 | | Crystal structure of CRISPR-associated protein | | Descriptor: | CRISPR-associated protein Cas1, GLYCEROL | | Authors: | Ka, D, Bae, E. | | Deposit date: | 2015-04-30 | | Release date: | 2016-01-13 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Streptococcus pyogenes Cas1 and Its Interaction with Csn2 in the Type II CRISPR-Cas System
Structure, 24, 2016
|
|
4QBI
 
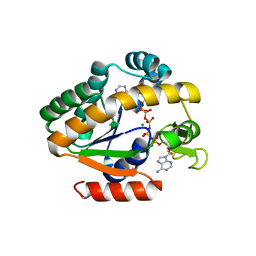 | | Crystal structure of a stable adenylate kinase variant AKlse6 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
4QBG
 
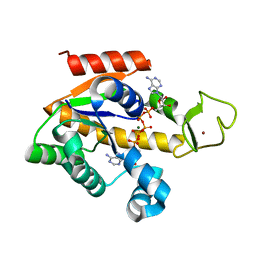 | | Crystal structure of a stable adenylate kinase variant AKlse4 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
4QBH
 
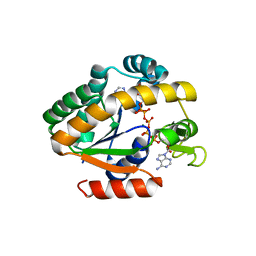 | | Crystal structure of a stable adenylate kinase variant AKlse5 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
4QBF
 
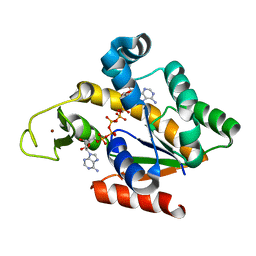 | | Crystal structure of a stable adenylate kinase variant AKlse2 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
5ZYF
 
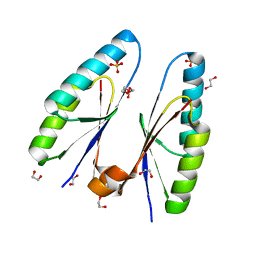 | | Crystal structure of Streptococcus pyogenes type II-A Cas2 | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endoribonuclease Cas2, SULFATE ION | | Authors: | Ka, D, Bae, E. | | Deposit date: | 2018-05-24 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2
Nucleic Acids Res., 46, 2018
|
|
5YHR
 
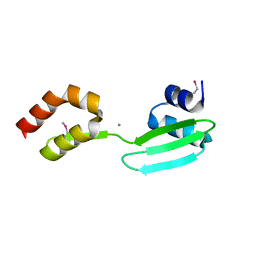 | | Crystal structure of the anti-CRISPR protein, AcrF2 | | Descriptor: | Anti-CRISPR protein 30, CALCIUM ION | | Authors: | Hong, S, Ka, D, Bae, E. | | Deposit date: | 2017-09-29 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | CRISPR RNA and anti-CRISPR protein binding to theXanthomonas albilineansCsy1-Csy2 heterodimer in the type I-F CRISPR-Cas system
J. Biol. Chem., 293, 2018
|
|
5X6I
 
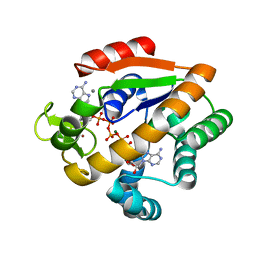 | | Crystal structure of B. subtilis adenylate kinase variant | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CALCIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mutational analyses of psychrophilic and mesophilic adenylate kinases highlight the role of hydrophobic interactions in protein thermal stability.
Struct Dyn., 6, 2019
|
|
5XZ2
 
 | | Crystal structure of adenylate kinase | | Descriptor: | Adenylate kinase isoenzyme 1, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, SULFATE ION | | Authors: | Moon, S, Bae, E, Kim, J. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analyses of adenylate kinases from Antarctic and tropical fishes for understanding cold adaptation of enzymes
Sci Rep, 7, 2017
|
|
5X6K
 
 | | Crystal structure of adenylate kinase | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, SULFATE ION, adenylate kinase isoenzyme 1 | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analyses of adenylate kinases from Antarctic and tropical fishes for understanding cold adaptation of enzymes
Sci Rep, 7, 2017
|
|
5Y69
 
 | | Crystal structure of the L52M mutant of AcrIIA1 | | Descriptor: | chain A | | Authors: | Ka, D, An, S.Y, Suh, J.Y, Bae, E. | | Deposit date: | 2017-08-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an anti-CRISPR protein, AcrIIA1.
Nucleic Acids Res., 46, 2018
|
|
5X6J
 
 | | Crystal structure of B. globisporus adenylate kinase variant | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mutational analyses of psychrophilic and mesophilic adenylate kinases highlight the role of hydrophobic interactions in protein thermal stability.
Struct Dyn., 6, 2019
|
|
