7S3F
 
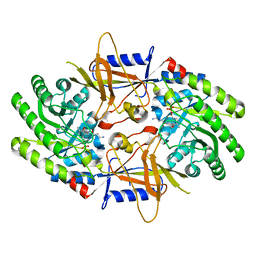 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3G
 
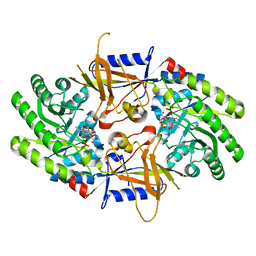 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
4GK7
 
 | | yeast 20S proteasome in complex with the Syringolin-Glidobactin chimera | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Groll, M, Stein, M.L, Bachmann, A. | | Deposit date: | 2012-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activity enhancement of the synthetic syrbactin proteasome inhibitor hybrid and biological evaluation in tumor cells.
Biochemistry, 51, 2012
|
|
7U6P
 
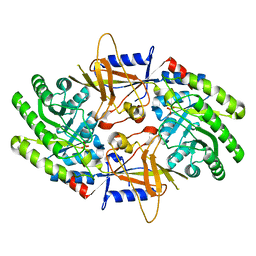 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R | | Descriptor: | Ornithine decarboxylase, PHOSPHATE ION | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
7U6U
 
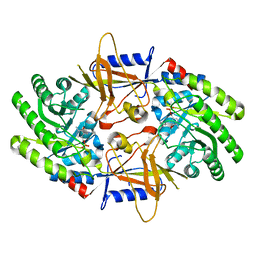 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R in complex with PLP | | Descriptor: | Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
8RYI
 
 | | Metformin hydrolase from Aminobacter niigataensis MD1 with urea in the active site | | Descriptor: | Agmatinase family protein, Arginase family protein, CALCIUM ION, ... | | Authors: | Fleming, J.R, Lutz, H, Bachmann, A, Mayans, O. | | Deposit date: | 2024-02-08 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Metformin hydrolase is a recently evolved, nickel-dependent, heteromeric ureohydrolase
To Be Published
|
|
