7XIO
 
 | | Crystal structure of TYR from Ralstonia | | Descriptor: | PHOSPHATE ION, Polyphenol oxidase | | Authors: | Sun, D.Y, Cui, P.P, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Zhang, J, Li, X, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of TYR from Ralstonia
To Be Published
|
|
7XFM
 
 | | Structure of nucleosome-AAG complex (A-53I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFC
 
 | | Structure of nucleosome-DI complex (-30I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFI
 
 | | Structure of nucleosome-DI complex (-50I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFH
 
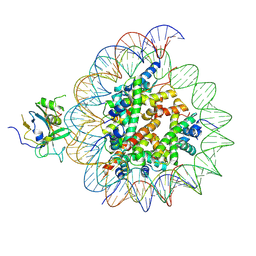 | | Structure of nucleosome-AAG complex (A-30I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XKK
 
 | | Human Cx36/GJD2 gap junction channel in detergents | | Descriptor: | Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XNP
 
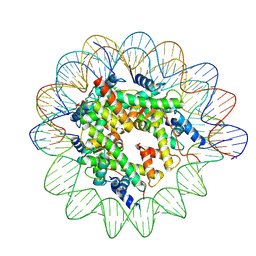 | | Structure of nucleosome-AAG complex (A-55I, post-catalytic state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7WUU
 
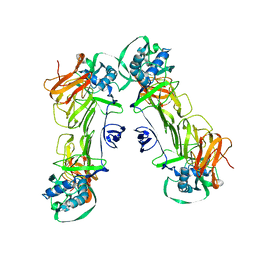 | | CryoEM structure of loose sNS1 tetramer | | Descriptor: | Core protein | | Authors: | Shu, B, Lok, S.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever.
Nat Commun, 13, 2022
|
|
7XL8
 
 | | Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7X4R
 
 | |
7XFJ
 
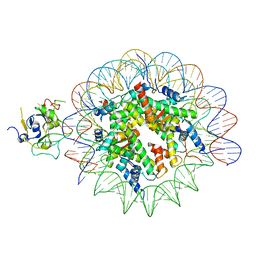 | | Structure of nucleosome-AAG complex (T-50I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
4S2M
 
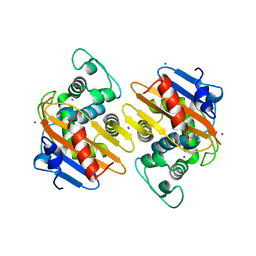 | | Crystal Structure of OXA-163 complexed with iodide in the active site | | Descriptor: | Beta-lactamase, IODIDE ION | | Authors: | Stojanoski, V, Hu, L, Palzkill, T.G, Prasad, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
7XW1
 
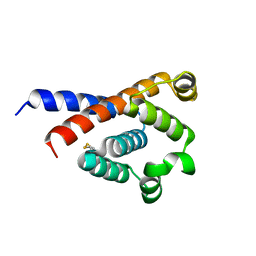 | |
7XXL
 
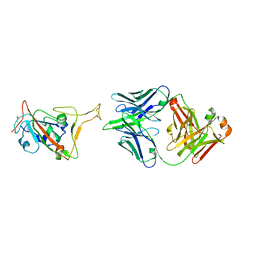 | | RBD in complex with Fab14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab14 heavy chain, Fab14 light chain, ... | | Authors: | Lin, J.Q, Tan, Y.J.E, Wu, B, Lescar, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-09-14 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
7Y45
 
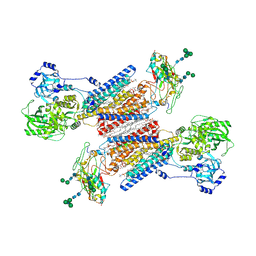 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
7YER
 
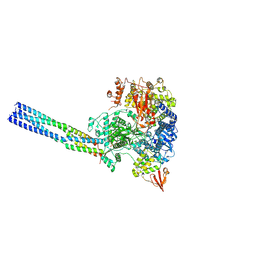 | | The structure of EBOV L-VP35 complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7Y46
 
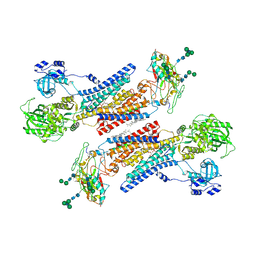 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state after addition of ATP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7YET
 
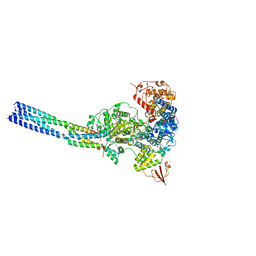 | | The structure of EBOV L-VP35 in complex with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Polymerase cofactor VP35, RNA-directed RNA polymerase L | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7YES
 
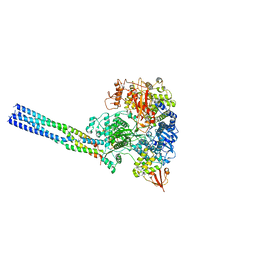 | |
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
