4EU2
 
 | | Crystal structure of 20s proteasome with novel inhibitor K-7174 | | 分子名称: | 1,4-bis[(4E)-5-(3,4,5-trimethoxyphenyl)pent-4-en-1-yl]-1,4-diazepane, Proteasome component C1, Proteasome component C11, ... | | 著者 | Kikuchi, J, Shibayama, N, Yamada, S, Wada, T, Nobuyoshi, M, Izumi, T, Akutsu, M, Kano, Y, Ohki, M, Sugiyama, K, Park, S.-Y, Furukawa, Y. | | 登録日 | 2012-04-25 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.509 Å) | | 主引用文献 | Homopiperazine derivatives as a novel class of proteasome inhibitors with a unique mode of proteasome binding.
Plos One, 8, 2013
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | 分子名称: | Beta-lactamase, CALCIUM ION, CESIUM ION | | 著者 | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | 登録日 | 2014-02-27 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WD7
 
 | | Type III polyketide synthase | | 分子名称: | COENZYME A, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Mori, T, Shimokawa, Y, Matsui, T, Kato, R, Sugio, S, Morita, H, Abe, I. | | 登録日 | 2013-06-10 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
1Q3F
 
 | | Uracil DNA glycosylase bound to a cationic 1-aza-2'-deoxyribose-containing DNA | | 分子名称: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*(NRI)P*AP*TP*CP*TP*T)-3', PHOSPHATE ION, ... | | 著者 | Bianchet, M.A, Seiple, L.A, Jiang, Y.L, Ichikawa, Y, Amzel, L.M, Stivers, J.T. | | 登録日 | 2003-07-29 | | 公開日 | 2004-03-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Electrostatic guidance of glycosyl cation migration along the reaction coordinate of uracil DNA glycosylase.
Biochemistry, 42, 2003
|
|
2P6M
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | 分子名称: | Alpha-ribazole-5'-phosphate phosphatase, GLYCEROL, SODIUM ION | | 著者 | Sugahara, M, Matsuura, Y, Morikawa, Y, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-19 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
4BH1
 
 | | H5 (tyTy) Influenza Virus Haemagglutinin in Complex with Avian Receptor Analogue 3'-SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | 登録日 | 2013-03-29 | | 公開日 | 2013-04-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BGZ
 
 | | Crystal Structure of H5 (tyTy) Influenza Virus Haemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ HA1, HEMAGGLUTININ, ... | | 著者 | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | 登録日 | 2013-03-29 | | 公開日 | 2013-04-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
4BH0
 
 | | H5 (tyTy) Influenza Virus Haemagglutinin in Complex with Human Receptor Analogue 6'-SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | 著者 | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | 登録日 | 2013-03-29 | | 公開日 | 2013-04-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
6KX7
 
 | | Crystal structure of mouse Cryptochrome 1 in complex with TH301 compound | | 分子名称: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-1 | | 著者 | Miller, S.A, Aikawa, Y, Hirota, T. | | 登録日 | 2019-09-10 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
1GIR
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | 分子名称: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | 登録日 | 2001-03-12 | | 公開日 | 2003-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
5GWA
 
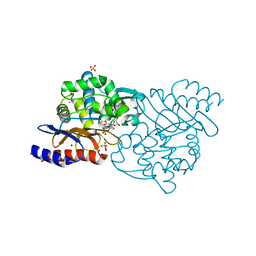 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with avibactam | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Wachino, J, Jin, W, Arakawa, Y. | | 登録日 | 2016-09-09 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
1GIQ
 
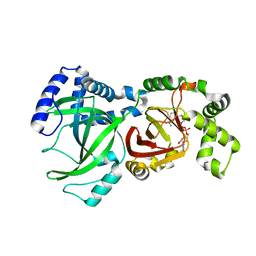 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | 著者 | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | 登録日 | 2001-03-12 | | 公開日 | 2003-01-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
2LCF
 
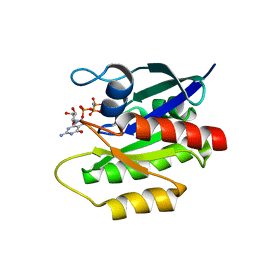 | | Solution structure of GppNHp-bound H-RasT35S mutant protein | | 分子名称: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Araki, M, Shima, F, Yoshikawa, Y, Muraoka, S, Ijiri, Y, Nagahara, Y, Shirono, T, Kataoka, T, Tamura, A. | | 登録日 | 2011-04-28 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers.
J.Biol.Chem., 286, 2011
|
|
2OWF
 
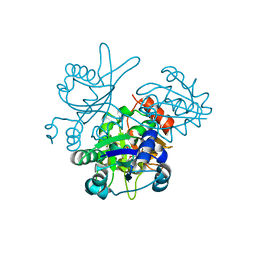 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | 著者 | Sugahara, M, Morikawa, Y, Matsuura, Y, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-02-16 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
5X9S
 
 | | Crystal structure of fully modified H-Ras-GppNHp | | 分子名称: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Matsumoto, S, Ke, H, Murashima, Y, Taniguchi-Tamura, H, Miyamoto, R, Yoshikawa, Y, Kumasaka, T, Mizohata, E, Edamatsu, H, Kataoka, T. | | 登録日 | 2017-03-09 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for intramolecular interaction of post-translationally modified H-RasGTP prepared by protein ligation
FEBS Lett., 591, 2017
|
|
2ZM0
 
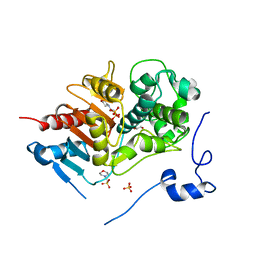 | | Structure of 6-aminohexanoate-dimer hydrolase, G181D/H266N/D370Y mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | 登録日 | 2008-04-10 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
3VGT
 
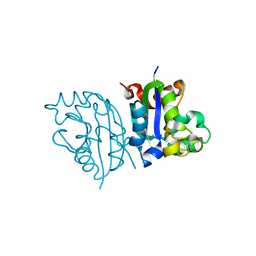 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | 分子名称: | Nucleoside diphosphate kinase | | 著者 | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | 登録日 | 2011-08-21 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | 分子名称: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2023-08-08 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7D0N
 
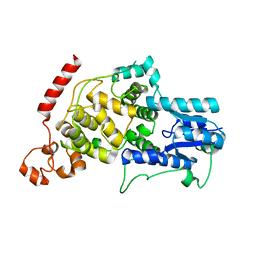 | |
7DLI
 
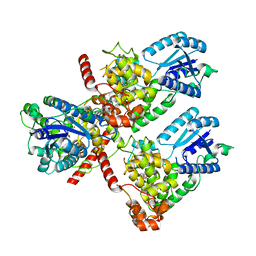 | | Crystal structure of mouse CRY1 in complex with KL001 compound | | 分子名称: | 1,2-ETHANEDIOL, Cryptochrome-1, N-[(2R)-3-carbazol-9-yl-2-oxidanyl-propyl]-N-(furan-2-ylmethyl)methanesulfonamide | | 著者 | Miller, S.A, Aikawa, Y, Hirota, T. | | 登録日 | 2020-11-27 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D0M
 
 | |
2ZJ9
 
 | | X-ray crystal structure of AmpC beta-Lactamase (AmpC(D)) from an Escherichia coli with a Tripeptide Deletion (Gly286 Ser287 Asp288) on the H10 Helix | | 分子名称: | AmpC, ISOPROPYL ALCOHOL, SODIUM ION | | 著者 | Yamaguchi, Y, Sato, G, Yamagata, Y, Wachino, J, Arakawa, Y, Kurosaki, H. | | 登録日 | 2008-02-29 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of AmpC beta-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5Z9W
 
 | | Ebola virus nucleoprotein-RNA complex | | 分子名称: | Ebolavirus nucleoprotein (residues 19-406), RNA (6-MER) | | 著者 | Sugita, Y, Matsunami, H, Kawaoka, Y, Noda, T, Wolf, M. | | 登録日 | 2018-02-05 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 angstrom resolution.
Nature, 563, 2018
|
|
7DQ5
 
 | | Crystal structure of HitB in complex with (S)-beta-phenylalanine sulfamoyladenosine | | 分子名称: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-phenyl-propanoyl]sulfamate | | 著者 | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | 登録日 | 2020-12-22 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7DQ6
 
 | | Crystal structure of HitB in complex with (S)-beta-3-Br-phenylalanine sulfamoyladenosine | | 分子名称: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-[(3S)-3-azanyl-3-(3-bromophenyl)propanoyl]sulfamate | | 著者 | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | 登録日 | 2020-12-22 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
