1XBB
 
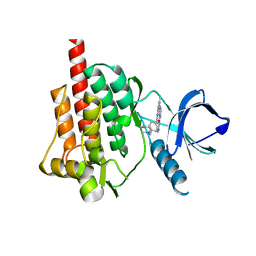 | | Crystal structure of the syk tyrosine kinase domain with Gleevec | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Tyrosine-protein kinase SYK | | Authors: | Nienaber, V.L, Atwell, S, Adams, J.M, Badger, J, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A Novel Mode of Gleevec Binding Is Revealed by the Structure of Spleen Tyrosine Kinase
J.Biol.Chem., 279, 2004
|
|
1XBC
 
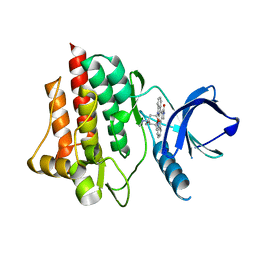 | | Crystal structure of the syk tyrosine kinase domain with Staurosporin | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase SYK | | Authors: | Badger, J, Atwell, S, Adams, J.M, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Nienaber, V.L, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase
J.Biol.Chem., 279, 2004
|
|
2IJQ
 
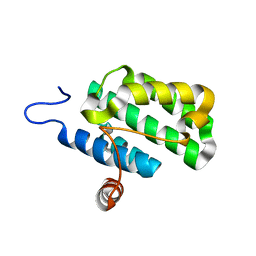 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
3JTY
 
 | | Crystal structure of a BenF-like porin from Pseudomonas fluorescens Pf-5 | | Descriptor: | BenF-like porin, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Sampathkumar, P, Lu, F, Zhao, X, Wasserman, S, Iuzuka, M, Bain, K, Rutter, M, Gheyi, T, Atwell, S, Luz, J, Gilmore, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution.
Proteins, 78, 2010
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3H9M
 
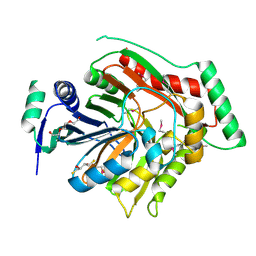 | | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, TRIETHYLENE GLYCOL, p-aminobenzoate synthetase, ... | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Do, J, Bain, K, Rutter, M, Gheyi, T, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii
To be Published
|
|
3HQC
 
 | | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sampathkumar, P, Romero, R, Wasserman, S, Do, J, Dickey, M, Bain, K, Gheyi, T, Klemke, R, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1)
To be Published
|
|
3E03
 
 | | Crystal structure of a putative dehydrogenase from Xanthomonas campestris | | Descriptor: | CALCIUM ION, Short chain dehydrogenase | | Authors: | Sampathkumar, P, Wasserman, S, Rutter, M, Hu, S, Bain, K, Rodgers, L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of a putative dehydrogenase from Xanthomonas campestris
To be Published
|
|
3NF5
 
 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
2OO6
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 | | Descriptor: | Putative L-alanine-DL-glutamate epimerase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Sridhar, V, Freeman, J, Smyth, L, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400
To be Published
|
|
2OO2
 
 | | Crystal structure of protein AF1782 from Archaeoglobus fulgidus, Pfam DUF357 | | Descriptor: | Hypothetical protein AF_1782 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Adams, J, Sridhar, V, Smyth, L, Freeman, J, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hypothetical AF_1782 protein from Archaeoglobus fulgidus
To be Published
|
|
4LLY
 
 | | Crystal structure of Pertuzumab Clambda Fab with variable and constant domain redesigns (VRD2 and CRD2) at 1.6A | | Descriptor: | GLYCEROL, MAGNESIUM ION, light chain Clambda, ... | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLQ
 
 | | Structure of redesigned IgG1 first constant and lambda domains (CH1:Clambda constant redesign 2 beta, CRD2b) at 1.42A | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, mutated CH1, mutated light chain Clambda | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLU
 
 | | Structure of Pertuzumab Fab with light chain Clambda at 2.16A | | Descriptor: | ACETATE ION, Light chain CLAMBDA, PERTUZUMAB FAB Heavy chain, ... | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLM
 
 | | Structure of redesigned IgG1 first constant and lambda domains (CH1:Clambda constant redesign 1, CRD1) at 1.75A | | Descriptor: | Ig gamma-1 chain C region, Ig lambda-2 chain C region | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLD
 
 | | Structure of wild-type IgG1 antibody heavy chain constant domain 1 and light chain lambda constant domain (IgG1 CH1:Clambda) at 1.19A | | Descriptor: | Ig gamma-1 chain C region, Ig lambda-2 chain C region | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLW
 
 | | Crystal structure of Pertuzumab Clambda Fab with variable domain redesign (VRD2) at 1.95A | | Descriptor: | SULFATE ION, light chain Clambda, mutated Pertuzumab Fab heavy chain | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
3GBW
 
 | | Crystal structure of the first PHR domain of the Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Klemke, R.L, Miller, S.A, Bain, K.T, Rutter, M.E, Tarun, G, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
3HWJ
 
 | | Crystal structure of the second PHR domain of Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Miller, S.A, Bain, K.T, Rutter, M.E, Gheyi, T, Klemke, R.L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
3P3D
 
 | | Crystal structure of the Nup53 RRM domain from Pichia guilliermondii | | Descriptor: | Nucleoporin 53 | | Authors: | Sampathkumar, P, Shawn, C, Bain, K, Gilmore, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the Nup53 RRM domain from Pichia guilliermondii
To be Published
|
|
3PVE
 
 | | Crystal structure of the G2 domain of Agrin from Mus Musculus | | Descriptor: | Agrin, Agrin protein | | Authors: | Sampathkumar, P, Do, J, Bain, K, Freeman, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the G2 domain of Agrin from Mus Musculus
To be Published
|
|
5CNI
 
 | | mGlu2 with Glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Clawson, D.K, Atwell, S, Monn, J.A. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
2HHL
 
 | | Crystal structure of the human small CTD phosphatase 3 isoform 1 | | Descriptor: | 12-TUNGSTOPHOSPHATE, CTD small phosphatase-like protein | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
