5KMZ
 
 | |
4P1Y
 
 | | Crystal structure of staphylococcal gamma-hemolysin prepore | | Descriptor: | Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2014-02-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins.
Nat Commun, 5, 2014
|
|
2LNC
 
 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
2Y8Y
 
 | |
2Y8W
 
 | |
2Y9H
 
 | |
7QCX
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, apo form | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
7QCY
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, complexed with RA-GEF2 peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
2FTD
 
 | | Crystal structure of Cathepsin K complexed with 7-Methyl-Substituted Azepan-3-one compound | | Descriptor: | Cathepsin K, N-[(1S)-1-({[(3S,4S,7R)-3-HYDROXY-7-METHYL-1-(PYRIDIN-2-YLSULFONYL)-2,3,4,7-TETRAHYDRO-1H-AZEPIN-4-YL]AMINO}CARBONYL)-3-METHYLBUTYL]-1-BENZOFURAN-2-CARBOXAMIDE | | Authors: | Yamashita, D.S, Baoguang, Z. | | Deposit date: | 2006-01-24 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure activity relationships of 5-, 6-, and 7-methyl-substituted azepan-3-one cathepsin K inhibitors.
J.Med.Chem., 49, 2006
|
|
1NC0
 
 | |
2O33
 
 | | Solution structure of U2 snRNA stem I from S. cerevisiae | | Descriptor: | U2 snRNA | | Authors: | Sashital, D.G, Venditti, V, Angers, C.A, Cornilescu, G, Butcher, S.E. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human.
Rna, 13, 2007
|
|
2O32
 
 | | Solution structure of U2 snRNA stem I from human, containing modified nucleotides | | Descriptor: | 5'-R(*(PSU)P*CP*UP*CP*(OMG)P*(OMG)P*CP*CP*(PSU)P*UP*UP*UP*(OMG)P*GP*CP*UP*AP*AP*(OMG)P*A)-3' | | Authors: | Sashital, D.G, Venditti, V, Angers, C.G, Cornilescu, G, Butcher, S.E. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human.
Rna, 13, 2007
|
|
6IIE
 
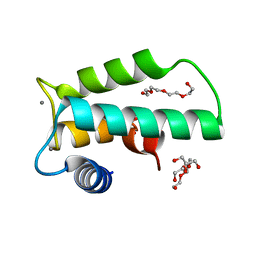 | | Crystal structure of human diacylglycerol kinase alpha EF-hand domains bound to Ca2+ | | Descriptor: | CALCIUM ION, Diacylglycerol kinase alpha, GLYCEROL, ... | | Authors: | Takahashi, D, Suzuki, K, Sakamoto, T, Iwamoto, T, Murata, T, Sakane, F. | | Deposit date: | 2018-10-04 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal structure and calcium-induced conformational changes of diacylglycerol kinase alpha EF-hand domains.
Protein Sci., 28, 2019
|
|
4AN8
 
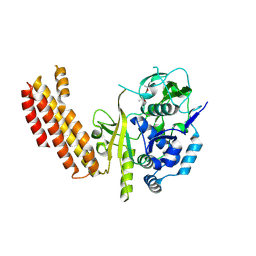 | |
4P1X
 
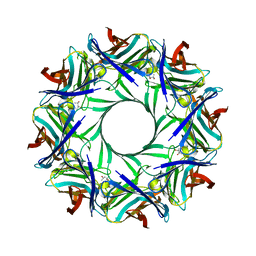 | | Crystal structure of staphylococcal LUK prepore | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-hemolysin component B, Gamma-hemolysin component C | | Authors: | Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2014-02-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins.
Nat Commun, 5, 2014
|
|
2M22
 
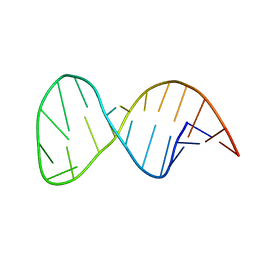 | | Solution structure of the helix II template boundary element from Tetrahymena telomerase RNA | | Descriptor: | 5'-R(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*UP*AP*AP*UP*AP*GP*AP*AP*CP*UP*GP*CP*C)-3' | | Authors: | Cash, D.D, Richards, R.J, Theimer, C.A, Finger, D.L, Feigon, J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the helix II template boundary element from Tetrahymena telomerase RNA
Nucleic Acids Res., 34, 2006
|
|
2M8K
 
 | | A pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo | | Descriptor: | RNA (48-MER) | | Authors: | Cash, D.D, Cohen, O, Kim, N, Shefer, K, Brown, Y, Ulyanov, N.B, Tzfati, Y, Feigon, J. | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3Q9T
 
 | | Crystal structure analysis of formate oxidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Doubayashi, D, Ootake, T, Maeda, Y, Oki, M, Tokunaga, Y, Sakurai, A, Nagaosa, Y, Mikami, B, Uchida, H. | | Deposit date: | 2011-01-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Formate oxidase, an enzyme of the glucose-methanol-choline oxidoreductase family, has a His-Arg pair and 8-formyl-FAD at the catalytic site.
Biosci.Biotechnol.Biochem., 75, 2011
|
|
1X8Q
 
 | | 0.85 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus in Complex with Water at pH 5.6 | | Descriptor: | Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
3FC6
 
 | | hRXRalpha & mLXRalpha with an indole Pharmacophore, SB786875 | | Descriptor: | Nr1h3 protein, RETINOIC ACID, Retinoic acid receptor RXR-alpha, ... | | Authors: | Washburn, D.G, Hoang, T.H, Campobasso, N, Smallwood, A, Parks, D.J, Webb, C.L, Frank, K, Nord, M, Duraiswami, C, Evans, C, Jaye, M, Thompson, S.K. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Synthesis and SAR of potent LXR agonists containing an indole pharmacophore.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7E1G
 
 | | Structure of MreB3 from Spiroplasma eriocheiris | | Descriptor: | Cell shape-determining protein MreB, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Takahashi, D, Miyata, M, Imada, K. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ATP-dependent polymerization dynamics of bacterial actin proteins involved in Spiroplasma swimming.
Open Biology, 12, 2022
|
|
7E1C
 
 | | Structure of MreB3 from Spiroplasma eriocheiris | | Descriptor: | ACETATE ION, CALCIUM ION, Cell shape-determining protein MreB | | Authors: | Takahashi, D, Miyata, M, Imada, K. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP-dependent polymerization dynamics of bacterial actin proteins involved in Spiroplasma swimming.
Open Biology, 12, 2022
|
|
1XHP
 
 | |
2A5X
 
 | | Crystal Structure of a Cross-linked Actin Dimer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Actin, alpha skeletal muscle, ... | | Authors: | Kudryashov, D.S, Sawaya, M.R, Adisetiyo, H, Norcross, T, Hegyi, G, Reisler, E, Yeates, T.O. | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of a cross-linked actin dimer suggests a detailed molecular interface in F-actin
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3TU5
 
 | | Actin complex with Gelsolin Segment 1 fused to Cobl segment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Kudryashov, D.S, Sawaya, M.R, Durer, Z.A.O. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural States and dynamics of the d-loop in actin.
Biophys.J., 103, 2012
|
|
